Band gap renormalization in diamond using one-shot method: Difference between revisions
| Line 314: | Line 314: | ||
<div class="toccolours mw-customtoggle-script">'''Click to show/''quasi_harm_4x4x4_diamond_create_pos_and_run_vasp.sh''</div> | <div class="toccolours mw-customtoggle-script">'''Click to show/''quasi_harm_4x4x4_diamond_create_pos_and_run_vasp.sh''</div> | ||
<div class="mw-collapsible mw-collapsed" id="mw-customcollapsible-script">{{:quasi_harm_4x4x4_diamond_create_pos_and_run_vasp}}</div> | <div class="mw-collapsible mw-collapsed" id="mw-customcollapsible-script">{{:quasi_harm_4x4x4_diamond_create_pos_and_run_vasp}}</div> | ||
This script creates 15 {{TAG|POSCAR}} files where the volume is varied in both directions in steps of 2 percent with respect to the starting volume. It also runs the necessary VASP calculations to obtain the dynamical matrix. To run this script please set your executable path and number of processors in the script and type: | |||
bash ./quasi_harm_4x4x4_diamond_create_pos_and_run_vasp.sh | |||
After that step one the free energy vs. volume curves need to be extracted for each temperature. To do this use the script: | |||
<div class="toccolours mw-customtoggle-script">'''Click to show/''quasi_harm_4x4x4_diamond_make_energy_vs_volume_plots.sh''</div> | |||
<div class="mw-collapsible mw-collapsed" id="mw-customcollapsible-script">{{:quasi_harm_4x4x4_diamond_make_energy_vs_volume_plots}}</div> | |||
To use this script please type: | |||
bash ./quasi_harm_4x4x4_diamond_make_energy_vs_volume_plots.sh | |||
<div class="toccolours mw-customtoggle-script">'''Click to show/''quasi_harm_4x4x4_diamond_obtain_fitting.sh''</div> | |||
<div class="mw-collapsible mw-collapsed" id="mw-customcollapsible-script">{{:quasi_harm_4x4x4_diamond_obtain_fitting}}</div> | |||
== References == | == References == | ||
Revision as of 07:56, 3 September 2020
Task
Calculating the zero-point renormalization and the temperature dependence of the band gap in diamond using a one-shot method.
Input
POSCAR
- Primitive cell (POSCAR.prim):
C_2_fcc
1.00000000000000
0.0000000000000000 1.783493 1.783493
1.783493 0.0000000000000000 1.783493
1.783493 1.783493 0.0000000000000000
C
2
Direct
0.0000000000000000 0.0000000000000000 0.0000000000000000
0.7500000000000000 0.7500000000000000 0.7500000000000000
- 4x4x4 super cell used in this calculation (POSCAR.4x4x4):
C_128_fcc
1.00000000000000
0.00000000 7.13397200 7.13397200
7.13397200 0.00000000 7.13397200
7.13397200 7.13397200 0.00000000
C
128
Direct
0.00000000 0.00000000 0.00000000
0.25000000 0.00000000 0.00000000
0.50000000 0.00000000 0.00000000
0.75000000 0.00000000 0.00000000
0.00000000 0.25000000 0.00000000
0.25000000 0.25000000 0.00000000
0.50000000 0.25000000 0.00000000
0.75000000 0.25000000 0.00000000
0.00000000 0.50000000 0.00000000
0.25000000 0.50000000 0.00000000
0.50000000 0.50000000 0.00000000
0.75000000 0.50000000 0.00000000
0.00000000 0.75000000 0.00000000
0.25000000 0.75000000 0.00000000
0.50000000 0.75000000 0.00000000
0.75000000 0.75000000 0.00000000
0.00000000 0.00000000 0.25000000
0.25000000 0.00000000 0.25000000
0.50000000 0.00000000 0.25000000
0.75000000 0.00000000 0.25000000
0.00000000 0.25000000 0.25000000
0.25000000 0.25000000 0.25000000
0.50000000 0.25000000 0.25000000
0.75000000 0.25000000 0.25000000
0.00000000 0.50000000 0.25000000
0.25000000 0.50000000 0.25000000
0.50000000 0.50000000 0.25000000
0.75000000 0.50000000 0.25000000
0.00000000 0.75000000 0.25000000
0.25000000 0.75000000 0.25000000
0.50000000 0.75000000 0.25000000
0.75000000 0.75000000 0.25000000
0.00000000 0.00000000 0.50000000
0.25000000 0.00000000 0.50000000
0.50000000 0.00000000 0.50000000
0.75000000 0.00000000 0.50000000
0.00000000 0.25000000 0.50000000
0.25000000 0.25000000 0.50000000
0.50000000 0.25000000 0.50000000
0.75000000 0.25000000 0.50000000
0.00000000 0.50000000 0.50000000
0.25000000 0.50000000 0.50000000
0.50000000 0.50000000 0.50000000
0.75000000 0.50000000 0.50000000
0.00000000 0.75000000 0.50000000
0.25000000 0.75000000 0.50000000
0.50000000 0.75000000 0.50000000
0.75000000 0.75000000 0.50000000
0.00000000 0.00000000 0.75000000
0.25000000 0.00000000 0.75000000
0.50000000 0.00000000 0.75000000
0.75000000 0.00000000 0.75000000
0.00000000 0.25000000 0.75000000
0.25000000 0.25000000 0.75000000
0.50000000 0.25000000 0.75000000
0.75000000 0.25000000 0.75000000
0.00000000 0.50000000 0.75000000
0.25000000 0.50000000 0.75000000
0.50000000 0.50000000 0.75000000
0.75000000 0.50000000 0.75000000
0.00000000 0.75000000 0.75000000
0.25000000 0.75000000 0.75000000
0.50000000 0.75000000 0.75000000
0.75000000 0.75000000 0.75000000
0.18750000 0.18750000 0.18750000
0.43750000 0.18750000 0.18750000
0.68750000 0.18750000 0.18750000
0.93750000 0.18750000 0.18750000
0.18750000 0.43750000 0.18750000
0.43750000 0.43750000 0.18750000
0.68750000 0.43750000 0.18750000
0.93750000 0.43750000 0.18750000
0.18750000 0.68750000 0.18750000
0.43750000 0.68750000 0.18750000
0.68750000 0.68750000 0.18750000
0.93750000 0.68750000 0.18750000
0.18750000 0.93750000 0.18750000
0.43750000 0.93750000 0.18750000
0.68750000 0.93750000 0.18750000
0.93750000 0.93750000 0.18750000
0.18750000 0.18750000 0.43750000
0.43750000 0.18750000 0.43750000
0.68750000 0.18750000 0.43750000
0.93750000 0.18750000 0.43750000
0.18750000 0.43750000 0.43750000
0.43750000 0.43750000 0.43750000
0.68750000 0.43750000 0.43750000
0.93750000 0.43750000 0.43750000
0.18750000 0.68750000 0.43750000
0.43750000 0.68750000 0.43750000
0.68750000 0.68750000 0.43750000
0.93750000 0.68750000 0.43750000
0.18750000 0.93750000 0.43750000
0.43750000 0.93750000 0.43750000
0.68750000 0.93750000 0.43750000
0.93750000 0.93750000 0.43750000
0.18750000 0.18750000 0.68750000
0.43750000 0.18750000 0.68750000
0.68750000 0.18750000 0.68750000
0.93750000 0.18750000 0.68750000
0.18750000 0.43750000 0.68750000
0.43750000 0.43750000 0.68750000
0.68750000 0.43750000 0.68750000
0.93750000 0.43750000 0.68750000
0.18750000 0.68750000 0.68750000
0.43750000 0.68750000 0.68750000
0.68750000 0.68750000 0.68750000
0.93750000 0.68750000 0.68750000
0.18750000 0.93750000 0.68750000
0.43750000 0.93750000 0.68750000
0.68750000 0.93750000 0.68750000
0.93750000 0.93750000 0.68750000
0.18750000 0.18750000 0.93750000
0.43750000 0.18750000 0.93750000
0.68750000 0.18750000 0.93750000
0.93750000 0.18750000 0.93750000
0.18750000 0.43750000 0.93750000
0.43750000 0.43750000 0.93750000
0.68750000 0.43750000 0.93750000
0.93750000 0.43750000 0.93750000
0.18750000 0.68750000 0.93750000
0.43750000 0.68750000 0.93750000
0.68750000 0.68750000 0.93750000
0.93750000 0.68750000 0.93750000
0.18750000 0.93750000 0.93750000
0.43750000 0.93750000 0.93750000
0.68750000 0.93750000 0.93750000
0.93750000 0.93750000 0.93750000
KPOINTS
K-Points 0 Gamma 1 1 1 0 0 0
- We only need a single k-point, since the convergence is done via the super-cell size.
INCAR
general: System = cd-C PREC = Accurate ALGO = FAST ISMEAR = 0 SIGMA = 0.1; IBRION = 6 PHON_LMC = .TRUE. PHON_NSTRUCT = 0 PHON_NTLIST = 1 PHON_TLIST = 0.0
- The tags with "PHON_" control the electron-phonon couplings (EPC). PHON_LMC enables electron-phonon coupling in the calculations using Monte Carlo sampling. By selecting PHON_NSTRUCT=0 a one-shot configuration (ZG configuration) is obtained. The tag PHON_NTLIST selects the number of temperatures for which the structure with the one shot calculation is obtained. This requires also the list of temperatures given by PHON_TLIST which have exact PHON_NTLIST number of elements.
- IBRION=6 is selected to obtain the eigenvectors and eigenvalues of the dynamical matrix at the Gamma point.
Calculation
This example will use a one-shot method, where only a single structure comprising the EPC is required for a given temperature.
The calculation consists of two steps:
- Obtain new "distorted" POSCAR file which contains the EPC. This calculation also contains the band gap of the original structure.
- Execute simple DFT calculation for the EPC structure to obtain the band gap.
- Extract ZPR as the difference between the band gaps from the two calculations.
Step 1 obtain structure containing EPC
To run the calculation POSCAR.4x4x4 needs to be copied to POSCAR and INCAR.init to INCAR.
Execute VASP. Execute the "Gamma" version of VASP since we only have a single k point in our calculations.
Copy the OUTCAR file to OUTCAR.init. It will be later used for the band gap of the "undistorted" structure.
The new POSCAR file containing the configurations according to the electron-phonon couplings is given as POSCAR.T=0..
Step 2 calculate electronic levels of structure containing EPC
Copy the file POSCAR.T=0. to POSCAR.
Delete (or comment out with #) all the lines in the INCAR file related to EPC so that it looks like the following:
System = cd-C PREC = Accurate ALGO = FAST ISMEAR = 0 SIGMA = 0.1;
Execute VASP.
Copy OUTCAR to OUTCAR.T=0..
Step 3 extract ZPR
We extract the band gap renormalization as
where and are the band gaps with and without EPC, respectively.
Since the 4x4x4 cell of cubic diamond as 3 degenerate bands at the valence band maximum and 6 degenerate bands at the conduction band minimum, they are averaged in the calculation of the band gap. This is necessary since the convergence with respect to cell size is drastically improved this way. Also all perturbation theory calculations in literature evaluate the band gap the same way, which ensures compatibility of the different calculational methods.
The band gaps are extracted from the previously saved files OUTCAR.init and OUTCAR.T=0. using the following script:
- REDIRECT Extract zpr
To use the script please type:
bash extract_zpr_cd_carbon.sh
The output of the script should look like the following:
The band gap (in eV) without zero-point vibrations is: 4.4049 The band gap (in eV) including zero-point vibrations is: 4.05102 The zero-point renormalization of the band gap (in eV) is: -0.353883
Step 4 better accuracy
The accuracy of the band gap renormalization depends dominantly on the size of the super cell, so this is the quantity that has to be usually converged in this type of calculation.
This example contains a POSCAR file for a 5x5x5 cell (POSCAR.5x5x5). After repeating all above steps with this POSCAR file the following results should be obtained:
The band gap (in eV) without zero-point vibrations is: 4.1421 The band gap (in eV) including zero-point vibrations is: 3.83717 The zero-point renormalization of the band gap (in eV) is: -0.304933
The interested user can try to further increase the cell size, by making a super cell from the primitive cell (POSCAR.prim) provided by this tutorial (how to build a super cell is for example covered here.
Step 6 temperature dependence of the band gap
Here the temperature dependence of the band gap with due to EPC is calculated. The input files are located in the directory TEMP_DEPENDENCE. Switch to this directory. Copy POSCAR.4x4x4 to POSCAR and INCAR.init to INCAR. The INCAR file contains following lines which are different from the calculation of the ZPR:
PHON_NTLIST = 8 PHON_TLIST = 0.0 100.0 200.0 300.0 400.0 500.0 600.0 700.0
After running the calculations several new POSCAR files are created. Run a standard VASP calculation for each of them to obtain the band gap or use the script run_temperature.sh provided with this calculation:
To run the script please edit the file and set your VASP executable path (vasp_exec) and the number of processors you are going to use (np). To run the calculation type:
bash ./run_temperature.sh
This step produces several OUTCAR files which can be analyzed using the script extract_temp.sh:
To run the script please type the following:
bash ./extract_temp.sh
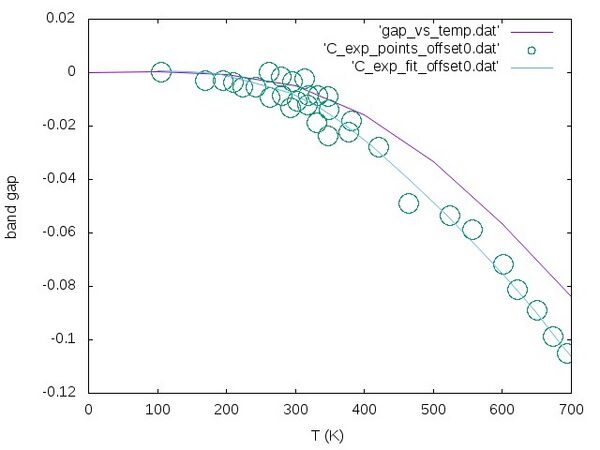
This script calculates the relative change of the band with respect to temperature (that means the offset is shifted to zero). The result ist outputted in gap_vs_temp.dat. To plot the data please type the following:
gnuplot -e "set terminal jpeg;set xlabel 'T (K)'; set ylabel 'band gap'; set style data lines; plot 'gap_vs_temp.dat', 'C_exp_points_offset0.dat' w circles, 'C_exp_fit_offset0.dat'" > gap_vs_temp.jpg
The resulting curve should look like the following:
The experimental data (blue lines and circles) are taken from reference [1].
Step 7 improving temperature dependence by adding volume effects
In the previous step we see that the experimental slope of the temperature dependence of the band gap is underestimated. To improve on that we will add also the volume dependence. The volume dependence is calculated from quasi harmonic calculations.
Go out of the directory ./TEMP_DEPENDENCE and go to the directory ./QUASI_HARMONIC. For the purpose of this tutorial the quasi harmonic calculations will be performed for the 4x4x4 cell but for exact calculations one needs to go to larger cell sizes until the results are converged.
There are three scripts in this directory to perform the quasi harmonic calculations:
- quasi_harm_4x4x4_diamond_create_pos_and_run_vasp.sh
- quasi_harm_4x4x4_diamond_make_energy_vs_volume_plots.sh
- quasi_harm_4x4x4_diamond_obtain_fitting.sh
For a quasi harmonic fit we need free energy vs. volume curves at different temperatures. Starting from the equilibrium volume we need to create POSCAR files at different volumes. The script to do this is given as:
This script creates 15 POSCAR files where the volume is varied in both directions in steps of 2 percent with respect to the starting volume. It also runs the necessary VASP calculations to obtain the dynamical matrix. To run this script please set your executable path and number of processors in the script and type:
bash ./quasi_harm_4x4x4_diamond_create_pos_and_run_vasp.sh
After that step one the free energy vs. volume curves need to be extracted for each temperature. To do this use the script:
To use this script please type:
bash ./quasi_harm_4x4x4_diamond_make_energy_vs_volume_plots.sh
References