XANES in Diamond: Difference between revisions
(improve language of Input section) |
(improve language of step 1 & 2) |
||
| Line 53: | Line 53: | ||
=== Step 1 build a supercell === | === Step 1 build a supercell === | ||
In the periodic boundary conditions of VASP, the core-hole interacts with its periodic replica so that we need sufficiently large super cells to reduce this spurious interaction. To this end, we employ successively larger cells until the spectrum shows no significant changes. For this tutorial, we illustrate the calculation of a core-hole using a <math>3\times3\times3</math> cell to allow for a reasonably fast calculation. However, for converged values one should use at least <math>4\times4\times4</math> super cell. | |||
The super cell can be obtained either | The super cell can be obtained by either taking the file POSCAR.3x3x3 provided with this tutorial or constructing the {{FILE|POSCAR}} file from the primitive cell using p4vasp, which is demonstrated below: | ||
*Open p4vasp by typing ''p4v'' on the terminal. | *Open p4vasp by typing ''p4v'' on the terminal. | ||
*Load the primitive cell by clicking on '''File'''→'''Load system''': | *Load the primitive cell by clicking on '''File'''→'''Load system''': | ||
| Line 65: | Line 65: | ||
=== Step 2 Prepare input files === | === Step 2 Prepare input files === | ||
The first few lines of the {{TAG|POSCAR}} file for the super cell should look like the following | The first few lines of the generated {{TAG|POSCAR}} file for the super cell should look like the following | ||
cubic diamond | cubic diamond | ||
3.567 | 3.567 | ||
+1.5000000000 +1.5000000000 +0.0000000000 | +1.5000000000 +1.5000000000 +0.0000000000 | ||
+0.0000000000 +1.5000000000 +1. | +0.0000000000 +1.5000000000 +1.5000000000 | ||
+1.5000000000 +0.0000000000 +1.5000000000 | +1.5000000000 +0.0000000000 +1.5000000000 | ||
54 | 54 | ||
| Line 77: | Line 77: | ||
+0.5000000000 +0.0000000000 +0.5000000000 | +0.5000000000 +0.0000000000 +0.5000000000 | ||
... | ... | ||
Here | Here, all the 54 atoms are of the same species (line 6). To distinguish between the atom with the core-hole and the rest, we treat one atom as a different species. Choosing the first atom, we replace the 54 in the 6th line with 1 and 53 and obtain the following {{TAG|POSCAR}} file | ||
cubic diamond | cubic diamond | ||
3.567 | 3.567 | ||
| Line 89: | Line 89: | ||
+0.5000000000 +0.0000000000 +0.5000000000 | +0.5000000000 +0.0000000000 +0.5000000000 | ||
... | ... | ||
In the {{TAG|INCAR}} file, we specify that the first species carries the core-hole by setting {{TAGBL|CLNT}}=1. We create a {{TAG|POTCAR}} file with the PAW/PS information for both species. Since both species are carbon this amounts simply to the concatenation of the {{TAG|POTCAR}} file for carbon | |||
cat POT_C POT_C > POTCAR | |||
We provide the resulting {{TAG|POTCAR}} file for this core-hole calculation in the tar file of this tutorial. | |||
The number of | To calculate accurate spectra, we need to include a sufficient number of conduction states. The required number of bands depends on the number of electrons in the system and the energy range of the spectrum. We can manually adjust the number of bands with the {{TAG|NBANDS}} variable. However, since the computation time increases drastically with the number of bands, selecting initially very large numbers is also not advisable. Hence, one has to increase the number of bands to find the optimum of computational effort and accuracy. In this tutorial, we use {{TAG|NBANDS}}=300. | ||
The | The other input variables in the {{TAG|INCAR}} file are described above. | ||
'''Mind''': The multiplicity of the species carrying the core hole has to be 1 otherwise the code will not work | '''Mind''': The multiplicity of the species carrying the core-hole has to be 1 otherwise the code will not work. Also mind that the selected species ({{TAGBL|CLNT}} in the {{TAG|INCAR}} file) is consistent with the order of the species specified in the {{TAG|POSCAR}} and {{TAG|POTCAR}} files. | ||
Revision as of 14:40, 7 December 2018
(UNDER CONSTRUCTION)
Important: This feature will be only available from VASP 6.0.
Task
Calculation of the XANES K-edge in diamond using the supercell core-hole method.
Input
POSCAR
cubic diamond 3.567 0.5 0.5 0.0 0.0 0.5 0.5 0.5 0.0 0.5 2 direct 0.0 0.0 0.0 0.25 0.25 0.25
Above, we show the POSCAR file for the primitive unit cell of diamond. Note that we will not use this structure as input for the calculation. Instead, we use it to construct a POSCAR file for a super cell actually used in the calculation.
INCAR
System = DIAMOND ALGO = FAST ISMEAR = 0; SIGMA = 0.1; ICORELEVEL = 2 CLNT = 1 CLN = 1 CLL = 0 CLZ = 1.0 CH_LSPEC = .TRUE. CH_SIGMA = 0.5 NBANDS = 300 LREAL = A
- To promote a core electron into the conduction bands and hence create the core-hole ICORELEVEL=2 has to be set. This corresponds to the final state approximation.
- CLNT=1 selects the first atom species in the POSCAR file.
- CLN=1 selects main quantum number 1 (hence K-edge).
- CLL=0 selects angular quantum number 0 (s).
- CLZ=1.0 selects the charge of the core hole. By setting this number to a fractional value we can mimic different screenings of the electrons. These non-integer values should only be used with caution, since this purely exploits error cancellation and does not correspond to a physically correct description of the screening.
- By setting CH_LSPEC=.TRUE., we enable the calculation of matrix elements between core and conduction states and the calculation of the core electron absorption spectrum.
- The broadening of the core electron absorption spectrum is controlled by the tag CH_SIGMA. Usually it is good practice to set this value low and broaden the spectrum in post processing.
- We have to set NBANDS to a larger value to consider enough conduction band states in the calculation.
- Since super cells are used the calculation of the projection operators in real space (LREAL=A) is much faster.
Calculation
Step 1 build a supercell
In the periodic boundary conditions of VASP, the core-hole interacts with its periodic replica so that we need sufficiently large super cells to reduce this spurious interaction. To this end, we employ successively larger cells until the spectrum shows no significant changes. For this tutorial, we illustrate the calculation of a core-hole using a cell to allow for a reasonably fast calculation. However, for converged values one should use at least super cell.
The super cell can be obtained by either taking the file POSCAR.3x3x3 provided with this tutorial or constructing the POSCAR file from the primitive cell using p4vasp, which is demonstrated below:
- Open p4vasp by typing p4v on the terminal.
- Load the primitive cell by clicking on File→Load system:
- Multiply cell in each direction (enter 3 for each direction) by clicking on Edit→Multiply Cell:
- Save new system by clicking on File→Save system as:
Step 2 Prepare input files
The first few lines of the generated POSCAR file for the super cell should look like the following
cubic diamond 3.567 +1.5000000000 +1.5000000000 +0.0000000000 +0.0000000000 +1.5000000000 +1.5000000000 +1.5000000000 +0.0000000000 +1.5000000000 54 Cartesian +0.0000000000 +0.0000000000 +0.0000000000 +0.2500000000 +0.2500000000 +0.2500000000 +0.5000000000 +0.0000000000 +0.5000000000 ...
Here, all the 54 atoms are of the same species (line 6). To distinguish between the atom with the core-hole and the rest, we treat one atom as a different species. Choosing the first atom, we replace the 54 in the 6th line with 1 and 53 and obtain the following POSCAR file
cubic diamond 3.567 +1.5000000000 +1.5000000000 +0.0000000000 +0.0000000000 +1.5000000000 +1.5000000000 +1.5000000000 +0.0000000000 +1.5000000000 1 53 Cartesian +0.0000000000 +0.0000000000 +0.0000000000 +0.2500000000 +0.2500000000 +0.2500000000 +0.5000000000 +0.0000000000 +0.5000000000 ...
In the INCAR file, we specify that the first species carries the core-hole by setting CLNT=1. We create a POTCAR file with the PAW/PS information for both species. Since both species are carbon this amounts simply to the concatenation of the POTCAR file for carbon
cat POT_C POT_C > POTCAR
We provide the resulting POTCAR file for this core-hole calculation in the tar file of this tutorial.
To calculate accurate spectra, we need to include a sufficient number of conduction states. The required number of bands depends on the number of electrons in the system and the energy range of the spectrum. We can manually adjust the number of bands with the NBANDS variable. However, since the computation time increases drastically with the number of bands, selecting initially very large numbers is also not advisable. Hence, one has to increase the number of bands to find the optimum of computational effort and accuracy. In this tutorial, we use NBANDS=300.
The other input variables in the INCAR file are described above.
Mind: The multiplicity of the species carrying the core-hole has to be 1 otherwise the code will not work. Also mind that the selected species (CLNT in the INCAR file) is consistent with the order of the species specified in the POSCAR and POTCAR files.
Step 3 Running Calculation
The SCF calculation with the core-hole and afterwards the calculation dielectric matrix (spectrum) is done in a single calculation. Usually the command to run the calculation looks like this: mpirun -np $np vasp_version, where $np corresponds to the number of processes and _version in the executable usually stands for std, gam, ncl namely standard, gamma-point only and non-collinear version, respectively. Since we have to use super cells in the calculation, which are large enough to minimize the interaction between core-holes on neighbouring cells, parallel execution with many computational cores is necessary to achieve good results within reasonable time. Hence for this example we use a cell, which gives sufficiently accurately results but more importantly finishes fast using only a few number of cores. For accurate results one should use at least a cell.
Step 4 Extraction of XAS Spectrum
To get the spectrum we want to plot the imaginary part of the frequency depedent dielectric function. This is written out in the OUTCAR file
frequency dependent IMAGINARY DIELECTRIC FUNCTION (independent particle, no local field effects) density-density
E(ev) X Y Z XY YZ ZX
--------------------------------------------------------------------------------------------------------------
243.589609 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000
243.677325 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000
243.765042 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000
...
Usually we are interested in the sum of all components of the dielectric matrix. You can either obtain this by your script or you use the script provided in this example. If you decide for the latter use the command perl ./plot_core_imdiel.pl. This will create the file CORE_DIELECTRIC_IMAG.dat which contains the summed up imaginary part of the dielectric matrix.
For this example we want to compare with experimental[1] and theoretical[2] XAS spectra from literature. They are provided in the files C_XAS_aligned_to_VASP.dat and C_PARATEC_aligned_to_VASP.dat. The literature calculations were obtained using the PARATEC code, which is also a PAW/Pseudopotential code. Both spectra are shifted to coincide with the VASP at the first peak. One can either plot these two spectra together with the VASP spectrum using his own prefered method of choice or using the script provided with this tutorial and invoking the command gnuplot gnuplot.script. One can also type plot.sh which does all the plotting steps together. The output should hopefully look like the this:
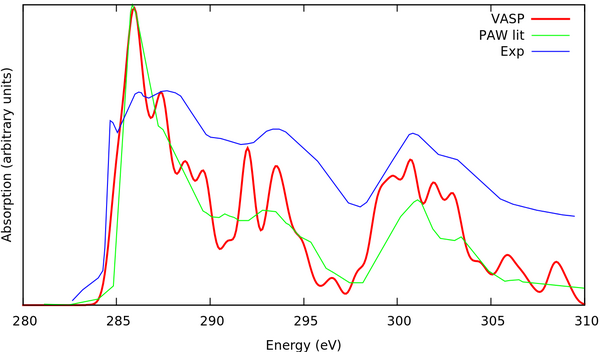
The experimental offset of the spectrum (absolute value of the first peak) is rather impossible to reproduce, since the calculated core energies are very different compared to experiment. Usually there is even a noticable deviation between calculations using different codes. It is accepted in literature to look at the relative peak positions in the spectra and the spectra can be scaled arbitrarily . In this example we scaled the experiment to VASP, since in this way the obtained results can be very easily compared to the experiment using a script. Usually one would either scale the first peak to 0 or would scale the calculated value to the experiment. Additionally the intensity of the spectrum can be scaled arbitrarily. So in this example we align the position and the height of the first peak for the calculations. The experiment is a little bit more tricky. It's a matter of taste what to consider as the first peak, but we decided that most likely the second peak corresponds to the first peak in the calculations and the first peak in experiment is a shoulder that is simply not pronounced in experiment.
Another important issue is the broadening. Many factors contribute to the experimental broadening depending totally on the experimental setup and hence it is impossible to reproduce the broadening exactly. We can choose a broadening that gives approximately the same width as experiment, but the choice of broadening parameters is rather arbitrary than systematic. For simplicity in this example we choose to use a 0.5 eV constant Lorentzian broadening (this is the only type of broadening built in for the calculation of XAS spectra in VASP). For more elaborate spectra we strongly advise users to choose a 0.05 eV broadening and apply the desired broadening in post-processing.
Apart from the obviously lower broadening width, we get quite reasonable agreement with theoretical literature calculations. We stress again that we used a cell in this example and that our results with respect to cell size are not fully converged. The interested user can repeat the calculations for the cell, which can be done in complete analogy to the cell (set NBANDS carefully since we have many more atoms in the cell). The files for this example are also given in the tar file.
Download
References
Back to the main page.