Machine learning force field: Theory
Here we present the theory for on-the-fly machine learning force fields. The theory will be presented in a very condensed manner and for a more detailed description of the methods, we refer the readers to Refs. [1], [2] and [3].
Introduction
Molecular dynamics is one of the most important methods for the determination of dynamic properties. The quality of the molecular dynamics simulation depends very strongly on the accuracy of the calculational method, but higher accuracy usually comes at the cost of increased computational demand. A very accurate method is ab initio molecular dynamics, where the interactions of atoms and electrons are calculated fully quantum mechanically from ab initio calculations (such as e.g. DFT). Unfortunately, this method is very often limited to small simulation cells and simulation times. One way to hugely speed up these calculations is by using force fields, which are parametrizations of the potential energy. These parametrizations can range from very simple functions with only empirical parameters, to very complex functions parametrized using thousands of ab initio calculations. Usually making accurate force fields manually is a very time-consuming task that needs tremendous expertise and know-how.
Another way to greatly reduce computational cost and required human intervention is by machine learning. Here, in the prediction of the target property, the method automatically interpolates between known training systems that were previously calculated ab initio. This way the generation of force fields is already significantly simplified compared to a classical force field which needs manual (or even empirical) adjustment of the parameters. Nevertheless, there is still the problem of how to choose the proper (minimal) training data. One very efficient and automatic way to solve that is to adapt on-the-fly learning. Here an MD calculation is used for the learning. During the run of the MD calculation, ab initio data is picked out and added to the training data. From the existing data, a force field is continuously built up. At each step, it is judged whether to make an ab initio calculation and possibly add the data to the force field or to use the force field for that step and actually skip learning for that step. Hence the name "on the fly" learning. The crucial point here is the probability model for the estimation of errors. This model is built up from newly sampled data. Hence, the more accurate the force field gets the less sampling is needed and the more expensive ab initio steps are skipped. This way not only the convergence of the force field can be controlled but also a very widespread scan through phase space for the training structures can be performed. The crucial point for on-the-fly machine learning which will be explained with the rest of the methodology in the following subsections is to be able to predict errors of the force field on a newly sampled structure without the necessity to perform an ab initio calculation on that structure.
Algorithms
On-the-fly machine-learning algorithm
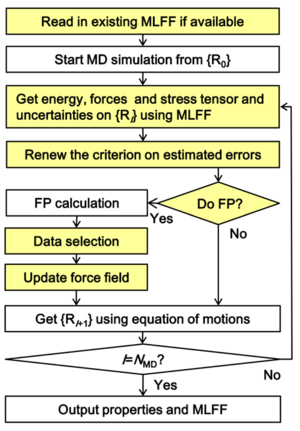
To obtain the machine-learning force field several structure datasets are required. A structure dataset defines the Bravais lattice and the atomic positions of the system and contains the total energy, the forces, and the stress tensor calculated by first principles. Given these structure datasets, the machine identifies local configurations around an atom to learn what force field is appropriate. The local configuration measures the radial and angular distribution of neighboring atoms around this given site and is captured in the so-called descriptors.
The on-the-fly force field generation scheme is given by the following steps (a flowchart of the algorithm is shown in Fig. 1):
- The machine predicts the energy, the forces, and the stress tensor and their uncertainties for a given structure using the existing force field.
- The machine decides whether to perform a first-principles calculation (proceed with step 3); otherwise we skip to step 5.
- Performing the first principles calculation, we obtain a new structure dataset that may improve the force field.
- If the number of newly collected structures reaches a certain threshold or if the predicted uncertainty becomes too large, the machine learns an improved force field by including the new structure datasets and local configurations.
- Atomic positions and velocities are updated by using the force field (if accurate enough) or the first principles calculation.
- If the desired total number of ionic steps (NSW) is reached we are done; otherwise proceed with step 1.
Sampling of training data and local reference configurations
We employ a learning scheme where structures are only added to the list of training structures when local reference configurations are picked for atoms that have an error in the force higher than a given threshold. So in the following, it is implied that whenever a new training structure is obtained, also local reference configurations from this structure are obtained.
Usually one can employ that the force field doesn't necessarily need to be retrained immediately at every step when a training structure with corresponding local configurations is added. Instead, one can also collect candidates and do the learning in a later step for all structures simultaneously (blocking). This way saving significant computational costs. Of course learning after every new configuration or after every block can have different results, but with not too large block sizes the difference should be small. The tag ML_MCONF_NEW sets the block size for learning. The force field is usually not updated at every molecular-dynamics step. The update happens under the following conditions:
- If there is no force field present, all atoms of a structure are sampled as local reference configurations and a force field is constructed.
- If the Bayesian error of the force for any atom is above the strict threshold set by ML_CDOUB[math]\displaystyle{ \times }[/math]ML_CTIFOR, the local reference configurations are sampled and a new force field is constructed.
- If the Bayesian error of the force for any atom is above the threshold ML_CTIFOR but below ML_CDOUB[math]\displaystyle{ \times }[/math]ML_CTIFOR, the structure is added to the list of new training structure candidates. Whenever the number of candidates is equal to ML_MCONF_NEW they are added to the entire set of training structures and the force field is updated. To avoid sampling too similar structures, the next step, from which training structures are allowed to be taken as candidates, is set by ML_NMDINT. All ab initio calculations within this distance are skipped if the Bayesian error for the force on all atoms is below ML_CDOUB[math]\displaystyle{ \times }[/math]ML_CTIFOR.
Threshold for error of forces
Training structures and their corresponding local configurations are only chosen if the error in the forces of any atom exceeds a chosen threshold. The initial threshold is set to the value provided by ML_CTIFOR (the unit is eV/Angstrom). The behavior of how the threshold is further controlled is given by ML_ICRITERIA. The following options are available:
- ML_ICRITERIA = 0: No update of initial value of ML_CTIFOR is done.
- ML_ICRITERIA = 1: Update of criteria using an average of the Bayesian errors of the forces from history (see description of the method below).
- ML_ICRITERIA = 2: Update of criteria using gliding average of Bayesian errors (probably more robust but not well tested).
Generally, it is recommended to automatically update the threshold ML_CTIFOR during machine learning. Details on how and when the update is performed are controlled by ML_CSLOPE, ML_CSIG and ML_MHIS.
Description of ML_ICRITERIA=1:
ML_CTIFOR is updated using the average Bayesian error in the previous steps. Specifically, it is set to
ML_CTIFOR = (average of the stored Bayesian errors) *(1.0 + ML_CX).
The number of entries in the history of the Bayesian errors is controlled by ML_MHIS. To avoid noisy data or an abrupt jump of the Bayesian error causing issues, the standard error of the history must be below the threshold ML_CSIG, for the update to take place. Furthermore, the slope of the stored data must be below the threshold ML_CSLOPE. In practice, the slope and the standard errors are at least to some extent correlated: often the standard error is proportional to ML_MHIS/3 times the slope or somewhat larger. We recommend to vary only ML_CSIG and keep ML_CSLOPE fixed to its default value.
Local energies
The potential energy [math]\displaystyle{ U }[/math] of a structure with [math]\displaystyle{ N_{a} }[/math] atoms is approximated as
[math]\displaystyle{ U = \sum\limits_{i=1}^{N_{\mathrm{a}}} U_{i}. }[/math]
The local energies [math]\displaystyle{ U_{i} }[/math] are functionals [math]\displaystyle{ U_{i}=F[\rho_{i}(\mathbf{r})] }[/math] of the probability density [math]\displaystyle{ \rho_{i} }[/math] to find another atom [math]\displaystyle{ j }[/math] at the position [math]\displaystyle{ \mathbf{r} }[/math] around the atom [math]\displaystyle{ i }[/math] within a cut-off radius [math]\displaystyle{ R_{\mathrm{cut}} }[/math] defined as
[math]\displaystyle{ \rho_{i}\left(\mathbf{r}\right) = \sum\limits_{j=1}^{N_{\mathrm{a}}} f_{\mathrm{cut}}\left(r_{ij}\right) g\left(\mathbf{r}-\mathbf{r}_{ij}\right), \qquad \qquad \qquad \qquad r_{ij}=|\mathbf{r}_{ij}|=|\mathbf{r}_{j}-\mathbf{r}_{i}|. }[/math]
Here [math]\displaystyle{ f_{\mathrm{cut}} }[/math] is a cut-off function that goes to zero for [math]\displaystyle{ r_{ij}\gt R_{\mathrm{cut}} }[/math] and [math]\displaystyle{ g\left(\mathbf{r}-\mathbf{r}_{ij}\right) }[/math] is a delta function.
The atom distribution can also be written as a sum of individual distributions:
[math]\displaystyle{ \rho_{i}\left(\mathbf{r}\right) = \sum\limits_{j \ne i}^{N_{\mathrm{a}}} \rho_{ij}\left(\mathbf{r}\right). }[/math]
Here [math]\displaystyle{ \rho_{ij}\left(\mathbf{r}\right) }[/math] describes the probability of find an atom [math]\displaystyle{ j }[/math] at a position [math]\displaystyle{ \mathbf{r} }[/math] with respect to atom [math]\displaystyle{ i }[/math].
Descriptors
Similar to the Smooth Overlap of Atomic Positions[4] (SOAP) method the delta function [math]\displaystyle{ g(\mathbf{r}) }[/math] is approximated as
[math]\displaystyle{ g\left(\mathbf{r}\right)=\frac{1}{\sigma_{\mathrm{atom}}\sqrt{2\pi}}\mathrm{exp}\left(-\frac{|\mathbf{r}|^{2}}{2\sigma_{\mathrm{atom}}^{2}}\right). }[/math]
Unfortunately [math]\displaystyle{ \rho_{i}\left(\mathbf{r}\right) }[/math] is not rotationally invariant. To deal with this problem intermediate functions or descriptors depending on [math]\displaystyle{ \rho_{i}\left(\mathbf{r}\right) }[/math] possessing rotational invariance are introduced:
Radial descriptor
This is the simplest descriptor which relies on the radial distribution function
[math]\displaystyle{ \rho_{i}^{(2)}\left(r\right) = \frac{1}{4\pi} \int \rho_{i}\left(r\hat{\mathbf{r}}\right) d\hat{\mathbf{r}}, }[/math]
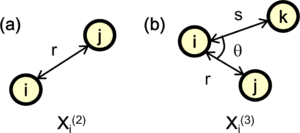
where [math]\displaystyle{ \hat{\mathbf{r}} }[/math] denotes the unit vector of the vector [math]\displaystyle{ \mathbf{r} }[/math] between atoms [math]\displaystyle{ i }[/math] and [math]\displaystyle{ j }[/math] [see Fig. 2 (a)]. The Radial descriptor can also be regarded as a two-body descriptor.
Angular descriptor
In most cases the radial descriptor is not enough to distinguish different probability densities [math]\displaystyle{ \rho_{i} }[/math], since two different [math]\displaystyle{ \rho_{i} }[/math] can yield the same [math]\displaystyle{ \rho_{i}^{(2)} }[/math]. To improve on this angular information between two radial descriptors is also incorporated within an angular descriptor
[math]\displaystyle{ \rho_{i}^{(3)}\left(r,s,\theta\right) = \iint d\hat{\mathbf{r}} d\hat{\mathbf{s}} \delta\left(\hat{\mathbf{r}}\cdot\hat{\mathbf{s}} - \mathrm{cos}\theta\right) \sum\limits_{j=1}^{N_{a}} \sum\limits_{k \ne j}^{N_{a}} \rho_{ik} \left(r\hat{\mathbf{r}}\right) \rho_{ij} \left(s\hat{\mathbf{s}}\right) }[/math]
where [math]\displaystyle{ \theta }[/math] denotes the angle between two vectors [math]\displaystyle{ \mathbf{r}_{ij} }[/math] and [math]\displaystyle{ \mathbf{r}_{ik} }[/math] [see Fig. 2 (b)]. The important difference of the function [math]\displaystyle{ \rho_{i}^{(3)} }[/math] compared to the angular distribution function (also called power spectrum within the Gaussian Approximation Potential) used in reference [5] is that no self interaction is included, where [math]\displaystyle{ j }[/math] and [math]\displaystyle{ k }[/math] have the same distance from [math]\displaystyle{ i }[/math] and the angle between the two is zero. It can be shown [3] that the self-interaction component is equivalent to the two body descriptors. This means that our angular descriptor is a pure angular descriptor, containing no two-body components and it cannot be expressed as linear combinations of the power spectrum. The advantage of this descriptor is that it enables us to separately control the effects of two- and three-body descriptors.
Basis set expansion and descriptors
The atomic probability density can be also expanded in terms of basis functions
[math]\displaystyle{ \rho_{i} \left( \mathbf{r} \right) = \sum\limits_{l=1}^{L_{\mathrm{max}}} \sum\limits_{m=-l}^{l} \sum\limits_{n=1}^{N^{l}_{ \mathrm{R}}} c_{nlm}^{i}\chi_{nl} \left( r \right) Y_{lm} \left( \hat{\mathbf{r}} \right), }[/math]
where [math]\displaystyle{ c_{nlm}^{i} }[/math], [math]\displaystyle{ \chi_{nl} }[/math] and [math]\displaystyle{ Y_{lm} }[/math] denote expansion coefficients, radial basis functions and spherical harmonics, respectively. The indices [math]\displaystyle{ n }[/math], [math]\displaystyle{ l }[/math] and [math]\displaystyle{ m }[/math] denote radial numbers, angular and magnetic quantum numbers, respectively.
By using the above equation the radial descriptor and angular descriptor can be written as
[math]\displaystyle{ \rho_{i}^{(2)}\left(r\right) = \frac{1}{\sqrt{4\pi}} \sum\limits_{n=1}^{N^{0}_{\mathrm{R}}} c_{n00}^{i} \chi_{n0}\left(r\right), }[/math]
and
[math]\displaystyle{ \rho_{i}^{(3)}\left(r,s,\theta\right) = \sum\limits_{l=1}^{L_{\mathrm{max}}} \sum\limits_{n=1}^{N^{l}_{\mathrm{R}}}\sum\limits_{\nu=1}^{N^{l}_{\mathrm{R}}} \sqrt{\frac{2l+1}{2}} p_{n\nu l}^{i}\chi_{nl}\left(r\right)\chi_{\nu l}\left(s\right)P_{l}\left(\mathrm{cos}\theta\right), }[/math]
where [math]\displaystyle{ \chi_{\nu l} }[/math] and [math]\displaystyle{ P_{l} }[/math] represent normalized spherical Bessel functions and Legendre polynomials of order [math]\displaystyle{ l }[/math], respectively.
The expansion coefficients for the angular descriptor can be converted to
[math]\displaystyle{ p_{n\nu l}^{i}=\sqrt{\frac{8\pi^{2}}{2l+1}} \sum\limits_{m=-l}^{l} \left[ c_{nlm}^{i} c_{\nu lm}^{i*} - \sum\limits_{j}^{N_{a}} c_{nlm}^{ij} c_{\nu lm}^{ij*}\right] , }[/math]
where [math]\displaystyle{ c_{nlm}^{ij} }[/math] denotes expansion coefficients of the distribution [math]\displaystyle{ \rho_{ij}(\mathbf{r}) }[/math] with respect to [math]\displaystyle{ \chi_{\nu l} }[/math] and [math]\displaystyle{ P_{l} }[/math]
[math]\displaystyle{ \rho_{ij}(\mathbf{r}) = \sum\limits_{l=1}^{L_{\mathrm{max}}} \sum\limits_{m=-l}^{l} \sum\limits_{n=1}^{N^{l}_{ \mathrm{R}}} c_{nlm}^{ij}\chi_{nl} \left( r \right) Y_{lm} \left( \hat{\mathbf{r}} \right) }[/math]
and
[math]\displaystyle{ c_{nlm}^{i} = \sum\limits_{j}^{N_{a}} c_{nlm}^{ij}. }[/math]
Angular filtering
In many cases [math]\displaystyle{ \chi_{nl} }[/math] is multiplied with an angular filtering function[6] [math]\displaystyle{ \eta }[/math] (ML_IAFILT2), which can noticably reduce the necessary basis set size without losing accuracy in the calculations
[math]\displaystyle{ \eta_{l,a_{\mathrm{FILT}}}=\frac{1}{1+a_{\mathrm{FILT}} [l (l+1)]^{2}} }[/math]
where [math]\displaystyle{ a_{\mathrm{FILT}} }[/math] (ML_AFILT2) is a parameter controlling the extent of the filtering. A larger value for the parameter should provide more filtering.
Reduced descriptors
A descriptor that reduces the number of descriptors with respect to the number of elements[7] is written as
[math]\displaystyle{ p_{n\nu l}^{iJ}=\sqrt{\frac{8\pi^{2}}{2l+1}} \sum\limits_{m=-l}^{l} c_{nlm}^{iJ} \sum\limits_{J'}c_{\nu lm}^{iJ'}. }[/math]
For more details on the effects of this descriptor please see following site: ML_DESC_TYPE.
Potential energy fitting
It is convenient to express the local potential energy of atom [math]\displaystyle{ i }[/math] in structure [math]\displaystyle{ \alpha }[/math] in terms of linear coefficients [math]\displaystyle{ w_{i_\mathrm{B}} }[/math] and a kernel [math]\displaystyle{ K \left(\mathbf{X}_{i},\mathbf{X}_{i_\mathrm{B}} \right) }[/math] as follows
[math]\displaystyle{ U_{i}^{\alpha} = \sum\limits_{i_\mathrm{B}=1}^{N_\mathrm{B}} w_{i_\mathrm{B}} K \left( \mathbf{X}_{i}^{\alpha},\mathbf{X}_{i_\mathrm{B}} \right) }[/math]
where [math]\displaystyle{ N_\mathrm{B} }[/math] is the basis set size. The kernel [math]\displaystyle{ K \left( \mathbf{X}_{i},\mathbf{X}_{i_\mathrm{B}}\right) }[/math] measures the similarity between a local configuration from the training set [math]\displaystyle{ \rho_{i}(\mathbf{r}) }[/math] and basis set [math]\displaystyle{ \rho_{i_\mathrm{B}}(\mathbf{r}) }[/math]. Using the radial and angular descriptors it is written as
[math]\displaystyle{ K \left(\mathbf{\hat{X}}_{i},\mathbf{\hat{X}}_{i_\mathrm{B}}\right) = \left[ \beta \mathbf{\hat{X}}_{i}^{(2)} \cdot \mathbf{\hat{X}}_{i_{\mathrm{B}}}^{(2)} + (1-\beta) \mathbf{\hat{X}}_{i}^{(3)} \cdot \mathbf{\hat{X}}_{i_{\mathrm{B}}}^{(3)} \right]^{\zeta}. }[/math]
Here the vectors [math]\displaystyle{ \mathbf{\hat{X}}_{i}^{(2)} }[/math] and [math]\displaystyle{ \mathbf{\hat{X}}_{i}^{(3)} }[/math] contain all coefficients [math]\displaystyle{ c_{n00}^{i} }[/math] and [math]\displaystyle{ p_{n\nu l}^{i} }[/math], respectively. The notation [math]\displaystyle{ \mathbf{\hat{X}} }[/math] indicates that it is a normalized vector. The parameter [math]\displaystyle{ \beta }[/math] (ML_W1) controls the weighting of the radial and angular terms, respectively. The parameter [math]\displaystyle{ \zeta }[/math] (ML_NHYP) controls the sharpness of the kernel and the order of the many-body interactions.
Normalization
The two and three body descriptors are combined to form a feature concatenated vector
[math]\displaystyle{ \mathbf{X}_{\mathrm{c}} = \begin{bmatrix} \sqrt{\beta} \mathbf{X}^{(2)} \\ \sqrt{(1-\beta)} \mathbf{X}^{(3)} \end{bmatrix}. }[/math]
[math]\displaystyle{ ||\mathbf{X}_{\mathrm{c}}|| }[/math] is used in the normalization of [math]\displaystyle{ \mathbf{X}^{(2)} }[/math] and [math]\displaystyle{ \mathbf{X}^{(3)} }[/math].
Matrix vector form of linear equations
Similarly to the energy [math]\displaystyle{ U_{i}^{\alpha} }[/math]the forces and the stress tensor are also described as linear functions of the coefficients [math]\displaystyle{ w_{i_\mathrm{B}} }[/math]. All three are fitted simultaneously which leads to the following matrix-vector form
[math]\displaystyle{ \mathbf{Y} = \mathbf{\Phi} \mathbf{w} }[/math]
where [math]\displaystyle{ \mathbf{Y} }[/math] is a super vector consisting of the sub vectors [math]\displaystyle{ \{\mathbf{y}^{\alpha}|\alpha=1,...,N_{\mathrm{st}}\} }[/math]. Here each [math]\displaystyle{ \mathbf{y}^{\alpha} }[/math] contains the first principle energies per atom, forces and stress tensors for each structure [math]\displaystyle{ \alpha }[/math]. [math]\displaystyle{ N_\mathrm{st} }[/math] denotes the total number of structures. The size of [math]\displaystyle{ \mathbf{Y} }[/math] is [math]\displaystyle{ N_{\mathrm{st}} \times (1+3N^{\alpha}_{a}+6) }[/math].
The matrix [math]\displaystyle{ \mathbf{\Phi} }[/math] is also called as design matrix[8]. The rows of this matrix are blocked for each structure [math]\displaystyle{ \alpha }[/math], where the first line of each block consists of the kernel used to calculate the energy. The subsequent [math]\displaystyle{ 3 N^{\alpha}_{a} }[/math] lines consist of the derivatives of the kernel with respect to the atomic coordinates used to calculate the forces. The final 6 lines within each structure consist of the derivatives of the kernel with respect to the unit cell coordinates used to calculate the stress tensor components. The overall size of [math]\displaystyle{ \mathbf{Y} }[/math] is [math]\displaystyle{ N_{\mathrm{st}} \times (1+3 N^{\alpha}_{a}+6)\times N_\mathrm{B} }[/math] looking like as follows
[math]\displaystyle{ \mathbf{\Phi} = \begin{bmatrix} \sum_{i} \frac{1}{N^{\alpha=1}_{\mathrm{a}}}K\left(\mathbf{X}^{\alpha=1}_{i},\mathbf{X}_{i_{\mathrm{B}}=1}\right) &\sum_{i} \frac{1}{N^{\alpha=1}_{\mathrm{a}}} K\left(\mathbf{X}^{\alpha=1}_{i},\mathbf{X}_{i_{\mathrm{B}}=2}\right) &\dots &\dots &\dots \\ \sum_{i} \nabla_{x_{1}} K\left(\mathbf{X}^{\alpha=1}_{i},\mathbf{X}_{i_{\mathrm{B}}=1}\right) &\sum_{i} \nabla_{x_{1}} K\left(\mathbf{X}^{\alpha=1}_{i},\mathbf{X}_{i_{\mathrm{B}}=2}\right) &\dots &\dots &\dots\\ \sum_{i} \nabla_{y_{1}} K\left(\mathbf{X}^{\alpha=1}_{i},\mathbf{X}_{i_{\mathrm{B}}=1}\right) &\sum_{i} \nabla_{y_{1}} K\left(\mathbf{X}^{\alpha=1}_{i},\mathbf{X}_{i_{\mathrm{B}}=2}\right) &\dots &\dots &\dots\\ \sum_{i} \nabla_{z_{1}} K\left(\mathbf{X}^{\alpha=1}_{i},\mathbf{X}_{i_{\mathrm{B}}=1}\right) &\sum_{i} \nabla_{z_{1}} K\left(\mathbf{X}^{\alpha=1}_{i},\mathbf{X}_{i_{\mathrm{B}}=2}\right) &\dots &\dots &\dots\\ \sum_{i} \nabla_{x_{2}} K\left(\mathbf{X}^{\alpha=1}_{i},\mathbf{X}_{i_{\mathrm{B}}=1}\right) &\sum_{i} \nabla_{x_{2}} K\left(\mathbf{X}^{\alpha=1}_{i},\mathbf{X}_{i_{\mathrm{B}}=2}\right) &\dots &\dots &\dots\\ \sum_{i} \nabla_{y_{2}} K\left(\mathbf{X}^{\alpha=1}_{i},\mathbf{X}_{i_{\mathrm{B}}=1}\right) &\sum_{i} \nabla_{y_{2}} K\left(\mathbf{X}^{\alpha=1}_{i},\mathbf{X}_{i_{\mathrm{B}}=2}\right) &\dots &\dots &\dots\\ \sum_{i} \nabla_{z_{2}} K\left(\mathbf{X}^{\alpha=1}_{i},\mathbf{X}_{i_{\mathrm{B}}=1}\right) &\sum_{i} \nabla_{z_{2}} K\left(\mathbf{X}^{\alpha=1}_{i},\mathbf{X}_{i_{\mathrm{B}}=2}\right) &\dots &\dots &\dots\\ \ldots &\ldots &\dots &\dots &\dots\\ \ldots &\ldots &\dots &\dots &\dots\\ \ldots &\ldots &\dots &\dots &\dots\\ \sum_{i} \frac{1}{N^{\alpha=2}_{\mathrm{a}}}K\left(\mathbf{X}^{\alpha=2}_{i},\mathbf{X}_{i_{\mathrm{B}}=1}\right) &\sum_{i} \frac{1}{N^{\alpha=2}_{\mathrm{a}}} K\left(\mathbf{X}^{\alpha=2}_{i},\mathbf{X}_{i_{\mathrm{B}}=2}\right) &\dots &\dots &\dots \\ \sum_{i} \nabla_{x_{1}} K\left(\mathbf{X}^{\alpha=2}_{i},\mathbf{X}_{i_{\mathrm{B}}=1}\right) &\sum_{i} \nabla_{x_{1}} K\left(\mathbf{X}^{\alpha=2}_{i},\mathbf{X}_{i_{\mathrm{B}}=2}\right) &\dots &\dots &\dots\\ \sum_{i} \nabla_{y_{1}} K\left(\mathbf{X}^{\alpha=2}_{i},\mathbf{X}_{i_{\mathrm{B}}=1}\right) &\sum_{i} \nabla_{y_{1}} K\left(\mathbf{X}^{\alpha=2}_{i},\mathbf{X}_{i_{\mathrm{B}}=2}\right) &\dots &\dots &\dots\\ \sum_{i} \nabla_{z_{1}} K\left(\mathbf{X}^{\alpha=2}_{i},\mathbf{X}_{i_{\mathrm{B}}=1}\right) &\sum_{i} \nabla_{z_{1}} K\left(\mathbf{X}^{\alpha=2}_{i},\mathbf{X}_{i_{\mathrm{B}}=2}\right) &\dots &\dots &\dots\\ \sum_{i} \nabla_{x_{2}} K\left(\mathbf{X}^{\alpha=2}_{i},\mathbf{X}_{i_{\mathrm{B}}=1}\right) &\sum_{i} \nabla_{x_{2}} K\left(\mathbf{X}^{\alpha=2}_{i},\mathbf{X}_{i_{\mathrm{B}}=2}\right) &\dots &\dots &\dots\\ \sum_{i} \nabla_{y_{2}} K\left(\mathbf{X}^{\alpha=2}_{i},\mathbf{X}_{i_{\mathrm{B}}=1}\right) &\sum_{i} \nabla_{y_{2}} K\left(\mathbf{X}^{\alpha=2}_{i},\mathbf{X}_{i_{\mathrm{B}}=2}\right) &\dots &\dots &\dots\\ \sum_{i} \nabla_{z_{2}} K\left(\dots\right) &\sum_{i} \nabla_{z_{2}} K\left(\dots\right) &\dots &\dots &\dots\\ \ldots &\ldots &\dots &\dots &\dots\end{bmatrix}. }[/math]
Bayesian linear regression
Ultimately for error prediction we want to get the maximized probability of observing a new structure [math]\displaystyle{ \mathbf{y} }[/math] on basis of the training set [math]\displaystyle{ \mathbf{Y} }[/math], which is denoted as [math]\displaystyle{ p \left( \mathbf{y} | \mathbf{Y} \right) }[/math]. For this we need to get from the error of the linear fitting coefficients [math]\displaystyle{ \mathbf{w} }[/math] in the reproduction of the training data [math]\displaystyle{ p\left( \mathbf{w} | \mathbf{Y} \right) }[/math] to [math]\displaystyle{ p \left( \mathbf{y} | \mathbf{Y} \right) }[/math] which is explained in the following.
First we obtain [math]\displaystyle{ p\left( \mathbf{w} | \mathbf{Y} \right) }[/math] from the Bayesian theorem
[math]\displaystyle{ p\left( \mathbf{w} | \mathbf{Y} \right) = \frac{p\left( \mathbf{Y} | \mathbf{w} \right) p\left( \mathbf{w} \right)}{p\left( \mathbf{Y} \right)} }[/math],
where we assume multivariate Gaussian distributions [math]\displaystyle{ \mathcal{N} }[/math] for the likelihood function
[math]\displaystyle{ p\left( \mathbf{Y} | \mathbf{w} \right) = \mathcal{N} \left(\mathbf{Y}| \mathbf{\Phi} \mathbf{w}, \sigma_{\mathrm{v}}^{2}\mathbf{I} \right) }[/math]
and the (conjugate) prior
[math]\displaystyle{ p\left( \mathbf{w} \right) = \mathcal{N} \left(\mathbf{w}| \mathbf{0},\sigma_{\mathrm{w}}^{2}\mathbf{I} \right). }[/math]
The parameters [math]\displaystyle{ \sigma_{\mathrm{w}}^{2} }[/math] and [math]\displaystyle{ \sigma_{\mathrm{v}}^{2} }[/math] need to be optimized to balance the accuracy and robustness of the force field (see below). The normalization is obtained by
[math]\displaystyle{ p\left( \mathbf{Y} \right) = \int p\left( \mathbf{Y} | \mathbf{w} \right) p\left( \mathbf{w} \right) d\mathbf{w}. }[/math]
Using the equations from above and the completing square method[8] [math]\displaystyle{ p\left( \mathbf{w} | \mathbf{Y} \right) }[/math] is obtained as follows
[math]\displaystyle{ p\left( \mathbf{w} | \mathbf{Y} \right) = \mathcal{N} \left(\mathbf{w}| \mathbf{\bar w},\mathbf{\Sigma} \right). }[/math]
Since the prior and the likelood function are described by multivariate Gaussian distributions the posterior [math]\displaystyle{ \mathcal{N} \left(\mathbf{w}| \mathbf{\bar w},\mathbf{\Sigma} \right) }[/math] describes a multivariate Gaussian written as
[math]\displaystyle{ \mathcal{N} \left(\mathbf{w}| \mathbf{\bar w},\mathbf{\Sigma} \right) = \frac{1}{\sqrt{ \left( 2 \pi \right)^{N_{\mathrm{B}}} \mathrm{det}\mathbf{\Sigma} }} \mathrm{exp} \left[ -\frac{ \left( \mathbf{w} - \mathbf{\bar w} \right)^{\mathrm{T}} \mathbf{\Sigma}^{-1} \left( \mathbf{w} - \mathbf{ \bar w} \right)}{2} \right] }[/math]
where we use the following definitions for the center of the Guassian distribution [math]\displaystyle{ \mathbf{\bar w} }[/math] and the covariance matrix [math]\displaystyle{ \mathbf{\Sigma} }[/math]
[math]\displaystyle{ \mathbf{\bar w} = s_{\mathrm{v}} \mathbf{\Sigma} \mathbf{\Phi}^{\mathrm{T}} \mathbf{Y}, }[/math]
[math]\displaystyle{ \mathbf{\Sigma}^{-1} =s_{\mathrm{w}} \mathbf{I} + s_{\mathrm{v}} \mathbf{\Phi}^{\mathrm{T}}\mathbf{\Phi}. }[/math]
Here we have used the following definitions
[math]\displaystyle{ s_{\mathrm{v}} = \frac{1}{\sigma_{\mathrm{v}}^{2}} }[/math]
and
[math]\displaystyle{ s_{\mathrm{w}} = \frac{1}{\sigma_{\mathrm{w}}^{2}}. }[/math]
Regression
We want to obtain the weights [math]\displaystyle{ \mathbf{\bar w} }[/math] for the linear equations
[math]\displaystyle{ \mathbf{Y} = \mathbf{\Phi} \mathbf{\bar w}. }[/math]
Solution via Inversion
By using directly the covariance matrix [math]\displaystyle{ \mathbf{\Sigma} }[/math] the equation from above
[math]\displaystyle{ \mathbf{\bar w} = s_{\mathrm{v}} \mathbf{\Sigma} \mathbf{\Phi}^{\mathrm{T}} \mathbf{Y}, }[/math]
can be solved straightforwardly. However, only [math]\displaystyle{ \mathbf{\Sigma}^{-1} }[/math] is directly accesible and [math]\displaystyle{ \mathbf{\Sigma} }[/math] must be obtained from [math]\displaystyle{ \mathbf{\Sigma}^{-1} }[/math] via inversion. This inversion is numerically unstable and leads to lower accuracy. Hence we don't use this method.
Solution via LU factorization
Here we directly use the invers of the covariance matrix [math]\displaystyle{ \mathbf{\Sigma}^{-1} }[/math] by solving the following equation for the weights [math]\displaystyle{ \mathbf{\bar w} }[/math]
[math]\displaystyle{ \mathbf{\Sigma}^{-1} \mathbf{\bar w} = s_{\mathrm{v}} \mathbf{\Phi}^{\mathrm{T}} \mathbf{Y}. }[/math]
For that [math]\displaystyle{ \mathbf{\Sigma}^{-1} }[/math] is decomposed into it's LU factorized components [math]\displaystyle{ \mathbf{P} * \mathbf{L} * \mathbf{U} }[/math]. After that the [math]\displaystyle{ \mathbf{L} * \mathbf{U} }[/math] factors are used to solve the previous linear equation for the weights [math]\displaystyle{ \mathbf{\bar w} }[/math]. This method is noticeably more accurate than the method via inversion of [math]\displaystyle{ \mathbf{\Sigma}^{-1} }[/math] while being on the same order of magnitude in terms of computational cost. Hence it is the method used in on-the-fly learning.
Solution via regularized SVD
The regression problem can also be solved by using the singular value decomposition to factorize [math]\displaystyle{ \mathbf{\Phi} }[/math] as
[math]\displaystyle{ \mathbf{\Phi}=\mathbf{U}\mathbf{\Lambda}\mathbf{V}^{T}. }[/math]
To add the regularization the diagonal matrix [math]\displaystyle{ \mathbf{\Lambda} }[/math] containing the singular values is rewritten as
[math]\displaystyle{ \tilde{\mathbf{\Lambda}}=\mathbf{\Lambda}+\frac{s_{\mathrm{w}}}{s_{\mathrm{v}}}\mathbf{\Lambda}^{-1}. }[/math]
By using the orthogonality of the left and right eigenvector matrices [math]\displaystyle{ \mathbf{U}^{T}\mathbf{U}=\mathbf{I}_{n} }[/math] and [math]\displaystyle{ \mathbf{V}\mathbf{V}^{T}=\mathbf{I}_{n} }[/math] the regression problem has the following solution
[math]\displaystyle{ \tilde{\mathbf{w}} = \mathbf{V}\tilde{\mathbf{\Lambda}}^{-1}\mathbf{U}^{T}\mathbf{Y}. }[/math]
Evidence approximation
Finally to get the best results and to prevent overfitting the parameters [math]\displaystyle{ \sigma_{\mathrm{v}}^{2} }[/math] and [math]\displaystyle{ \sigma_{\mathrm{w}}^{2} }[/math] have to be optimized. To achieve this, we use the evidence approximation[9][10][11] (also called as empirical bayes, 2 maximum likelihood or generalized maximum likelihood), which maximizes the evidence function (also called model evidence) defined as
[math]\displaystyle{ p\left(\mathbf{Y}|\sigma_{\mathrm{v}}^{2},\sigma_{\mathrm{w}}^{2}\right) =\left(\frac{1}{\sqrt{2\pi\sigma_{\mathrm{v}}^{2}}}\right)^{M} \left(\frac{1}{\sqrt{2\pi\sigma_{\mathrm{w}}^{2}}}\right)^{N_{\mathrm{B}}} \int \mathrm{exp}\left[-E\left( \mathbf{w} \right) \right] d\mathbf{w}, }[/math]
[math]\displaystyle{ E\left( \mathbf{w} \right) = \frac{s_{\mathrm{v}}}{2} || \mathbf{\Phi}\mathbf{w}-\mathbf{Y}||^{2} + \frac{s_{\mathrm{w}}}{ 2}||\mathbf{w}||^{2}. }[/math]
The function [math]\displaystyle{ p\left(\mathbf{Y}|\sigma_{\mathrm{v}}^{2},\sigma_{\mathrm{w}}^{2}\right) }[/math] corresponds to the probability that the regression model with parameters [math]\displaystyle{ \sigma_{\mathrm{v}}^{2} }[/math] and [math]\displaystyle{ \sigma_{\mathrm{w}}^{2} }[/math] provides the reference data [math]\displaystyle{ \mathbf{Y} }[/math]. Hence the best fit is optimized by maximizing this probability. The maximization is carried out by simultaneously solving the following equations
[math]\displaystyle{ s_{\mathrm{w}}=\frac{\gamma}{|\mathbf{\bar{w}}|^{2}}, }[/math]
[math]\displaystyle{ s_{\mathrm{v}}=\frac{M-\gamma}{|\mathbf{T}-\mathbf{\phi}\mathbf{\bar{w}}|^{2}}, }[/math]
[math]\displaystyle{ \gamma=\sum\limits_{k=1}^{N_{\mathrm{B}}} \frac{\lambda_{k}}{\lambda_{k}+s_{\mathrm{w}}} }[/math]
where [math]\displaystyle{ \lambda_{k} }[/math] are the eigenvalues of the matrix [math]\displaystyle{ s_{\mathrm{v}}\mathbf{\Phi}^{\mathrm{T}}\mathbf{\Phi} }[/math].
The evidence approximation can be done for any of the above-described regression methods, but we combine it only with the solutions from LU factorization since solutions via SVD are calculationally too expensive to be carried out multiple times.
Error estimation
Error estimates from Bayesian linear regression
By using the relation
[math]\displaystyle{ p \left( \mathbf{y} | \mathbf{Y} \right) = \int p \left( \mathbf{y} | \mathbf{w} \right) p \left( \mathbf{w} | \mathbf{Y} \right) d\mathbf{w} }[/math]
and the completing square method[8] the distribution of [math]\displaystyle{ p \left( \mathbf{y} | \mathbf{Y} \right) }[/math] is written as
[math]\displaystyle{ p \left( \mathbf{y} | \mathbf{Y} \right) = \mathcal{N} \left( \mathbf{\phi}\mathbf{\bar w}, \mathbf{\sigma} \right), }[/math]
[math]\displaystyle{ \mathbf{\sigma}=\frac{1}{s_{\mathrm{v}}}\mathbf{I}+\mathbf{\phi}^{\mathrm{T}}\mathbf{\Sigma}\mathbf{\phi}. }[/math]
Here [math]\displaystyle{ \mathbf{\phi} }[/math] is similar to [math]\displaystyle{ \mathbf{\Phi} }[/math] but only for the new structure [math]\displaystyle{ \mathbf{y} }[/math].
The mean vector [math]\displaystyle{ \mathbf{\phi}\mathbf{\bar w} }[/math] contains the results of the predictions on the dimensionless energy per atom, forces, and stress tensor. The diagonal elements of [math]\displaystyle{ \mathbf{\sigma} }[/math], which correspond to the variances of the predicted results, are used as the uncertainty in the prediction.
Spilling factor
The spilling factor[2][12] [math]\displaystyle{ s_{i} }[/math] is a measure of the overlap (or similarity) of a given structural environment on an atom [math]\displaystyle{ \mathbf{X}_{i} }[/math] with the local reference configurations [math]\displaystyle{ \mathbf{X}_{i_{\mathrm{B}}} }[/math] written as
[math]\displaystyle{ s_{i}= 1 - \frac{ \sum\limits_{i_{\mathrm{B}}=1}^{N_{\mathrm{B}}} \sum\limits_{i'_{\mathrm{B}}=1}^{N_{\mathrm{B}}} K(\mathbf{X}_{i},\mathbf{X}_{i_{\mathrm{B}}}) K^{-1}(\mathbf{X}_{i_{\mathrm{B}}}, \mathbf{X}_{i'_{\mathrm{B}}}) K(\mathbf{X}_{i'_{\mathrm{B}}},\mathbf{X}_{i}) } { K(\mathbf{X}_{i},\mathbf{X}_{i}) }. }[/math]
If [math]\displaystyle{ \mathbf{X}_{i} }[/math] is fully overlapping with any of the local reference configurations then the second term on the right hand side of the above equation becomes 1 and [math]\displaystyle{ s = 0 }[/math]. If [math]\displaystyle{ \mathbf{X}_{i} }[/math] has no similarities with any of the local reference configurations the second term on the right hand side of the above equations becomes 0 and [math]\displaystyle{ s = 1 }[/math].
Sparsification
Within the machine learning force field methods the sparsification of local reference configurations and the angular descriptors is supported. The sparsification of local reference configurations is by default used and the extent is mainly controlled by ML_EPS_LOW. This is procedure is important to avoid overcompleteness and to dampen the acquisition of new configurations in special cases. The sparsification of angular descriptors is by default not used and should be used very cautiously and only if it's necessary. The description of the usage of this feature is given in ML_LSPARSDES.
Sparsification of local reference configurations
We start by defining the similarity kernel (or Gram matrix) for the local configurations with each other
[math]\displaystyle{ \mathbf{K}_{i_{B},j_{B}}=\mathbf{K} \left(\mathbf{X}_{i_B},\mathbf{X}_{j_B}\right). }[/math]
The CUR algorithm starts out from the diagonalization of this matrix
[math]\displaystyle{ \mathbf{U}^{T}\mathbf{K} \mathbf{U}= \mathbf{L} = \textrm{diag}(l_{1},l_{2},...,l_{N_{B}}), }[/math]
where [math]\displaystyle{ \mathbf{U} }[/math] is the matrix of the eigenvectors [math]\displaystyle{ \mathbf{u_j} }[/math]
[math]\displaystyle{ \mathbf{U} = (\mathbf{u}_{1},\mathbf{u}_{2},...,\mathbf{u}_{N_{B}}), \qquad \mathbf{u}_{j} = \left( \begin{array}{c} u_{1j} \\ u_{2j} \\... \\ u_{N_{B}j} \end{array} \right) }[/math]
In contrast to the original CUR algorithm[13] that was developed to efficiently select a few significant columns of the matrix [math]\displaystyle{ \mathbf{K} }[/math], we search for (few) insignificant local configurations and remove them. We dispose of the columns of [math]\displaystyle{ \mathbf{K} }[/math] that are correlated with the [math]\displaystyle{ N_{\mathrm{low}} }[/math] eigenvectors [math]\displaystyle{ u_{\chi} }[/math] with the smallest eigenvalues [math]\displaystyle{ l_{\chi} }[/math]. The correlation is measured by the statistical leverage scoring measured for each column [math]\displaystyle{ j }[/math] of [math]\displaystyle{ \mathbf{K} }[/math] as
[math]\displaystyle{ \omega_{j}= \frac{1}{N_{\mathrm{low}}} \sum\limits_{\chi=1}^{N_{B}} \gamma_{\chi,j}, }[/math]
[math]\displaystyle{ \gamma_{\chi,j} = \left\{ \begin{array}{cl} u_{j\chi}^{2} &\mathrm{if} \,\, l_{\chi} \gt \epsilon_{\mathrm{low}} \\ 0 &\mathrm{otherwise} \end{array} \right. , }[/math]
where [math]\displaystyle{ \epsilon_{\mathrm{low}} }[/math] (see also ML_EPS_LOW) is the threshold for the lowest eigenvalues. One can prove (using the orthogonality of the eigenvectors and their completeness relation) that this is equivalent to the usual CUR leverage scoring algorithm, i.e. removing the least significant columns will result in those columns that are most strongly "correlated" to the largest eigenvalues.
Sparsification of angular descriptors
The sparsification of the angular descriptors is done in a similar manner as for the local reference configurations. We start by defining the [math]\displaystyle{ N_{D} \times N_{D} }[/math] square matrix
[math]\displaystyle{ \mathbf{A}=\mathbf{X}\mathbf{X}^{T}. }[/math]
Here [math]\displaystyle{ N_{D} }[/math] denotes the number of angular descriptors given by
[math]\displaystyle{ N_{D} = \sum\limits_{l=1}^{L_{\mathrm{max}}} \frac{1}{2} N_{R}^{l} \left( N_{R}^{l} + 1 \right) \left( L_{\mathrm{max}} + 1 \right). }[/math]
In this equation the symmetry of the descriptors [math]\displaystyle{ \rho_{n \nu l}= \rho_{\nu n l} }[/math] is already taken into account. The matrix [math]\displaystyle{ \mathbf{X} }[/math] is constructed from the vectors of the angular descriptors [math]\displaystyle{ \left(\mathbf{x}_{1}^{(3)},\mathbf{x}_{2}^{(3)},...,\mathbf{x}_{N_{B}}^{(3)} \right) }[/math]. The matrix [math]\displaystyle{ \mathbf{X} }[/math] has dimension [math]\displaystyle{ N_{D} \times N_{B} }[/math] and the matrix product is done over the [math]\displaystyle{ N_{B} }[/math] elements of the local configurations.
In analogy to the local configurations the [math]\displaystyle{ j }[/math]th element of the matrix [math]\displaystyle{ \mathbf{A} }[/math] is written as
[math]\displaystyle{ \mathbf{a}_{j} = \sum\limits_{\chi=1}^{N_{D}} (u_{j\chi}l_{\chi})\mathbf{u_{\chi}}. }[/math]
In contrast to the sparsification of the local configuration and more in line with the original CUR method, the columns of matrix [math]\displaystyle{ \mathbf{A} }[/math] are kept when they are strongly correlated to the [math]\displaystyle{ k }[/math] (see also ML_NRANK_SPARSDES) eigenvectors [math]\displaystyle{ u_{\chi} }[/math] which have the largest eigenvalues [math]\displaystyle{ l_{\chi} }[/math]. The correlation of the eigenvalues is then measured via a leverage scoring
[math]\displaystyle{ \omega_{j} = \frac{1}{k} \sum\limits_{\chi=1}^{k} u_{j\chi}^{2}. }[/math]
From the leverage scorings, the ones with the highest values are selected until the ratio of the selected descriptors to the total number of descriptors becomes a previously selected value [math]\displaystyle{ x_{\mathrm{spars}} }[/math] (see also ML_RDES_SPARSDES).
References
- ↑ R. Jinnouchi, J. Lahnsteiner, F. Karsai, G. Kresse, and M. Bokdam, Phys. Rev. Lett. 122, 225701 (2019).
- ↑ a b R. Jinnouchi, F. Karsai, and G. Kresse, Phys. Rev. B 100, 014105 (2019).
- ↑ a b R. Jinnouchi, F. Karsai, C. Verdi, R. Asahi, and G. Kresse, J. Chem. Phys. 152, 234102 (2020).
- ↑ A. P. Bartók, R. Kondor, and G. Csányi, Phys. Rev. B 87, 184115 (2013).
- ↑ A. P. Bartók, M.C. Payne, R. Kondor, and G. Csányi, Phys. Rev. Lett 104, 136403 (2010).
- ↑ J. P. Boyd, Chebyshev and Fourier Spectral Methods (Dover Publications, New York, 2000).
- ↑ J. P. Darby, J. R. Kermode, and G. Csanyi, Compressing local atomic neighbourhood descriptors, New Phys. J. 8, 166 (2022).
- ↑ a b c C. M. Bishop, Pattern Recognition and Machine Learning, (New York: Springer), (2006).
- ↑ S.F. Gull and J. Skilling, Maximum Entropy Bayesian Methods, Fundam. Theor. Phys., 28th ed. (Springer, Dordrecht, 1989).
- ↑ D. J. C. Mackay, Neural Computation 4, 415 (1992).
- ↑ R. Jinnouchi and R. Asahi, J. Phys. Chem. Lett. 8, 4279 (2017).
- ↑ K. Miwa and H. Ohno, Phys. Rev. B 94, 184109 (2016).
- ↑ M. W. Mahoney and P. Drineas, Proc. Natl. Acad. Sci. USA 106, 697 (2009).
