Best practices for machine-learned force fields
Using the machine-learning–force-fields method, VASP can construct force fields based on ab-initio simulations. There are many aspects to carefully consider while constructing, testing, retraining, and applying a force field. Here, we list some best practices but bare in mind that the list is incomplete and the method has not been applied to a large number of systems. Therefore, we recommend using the usual rigorous checking that is necessary for any research project.
General advices
On-the-fly learning involving molecular dynamics is significantly more involved than single-point electronic calculations since it depends on the following:
- ) Electronic calculations for each training point.
- ) Molecular dynamics controlling the correct learning conditions.
- ) Machine learning controlling the sampling.
Please consider the following general advices:
- ) Provide a minimal INCAR file, where only the tag-value pairs are provided that different from the default.
- ) Order the tags in the INCAR to groups that belong together via their functionality (electronic convergence, molecular-dynamics, output options, machine learning, etc.).
- ) Use a random seed (RANDOM_SEED) when molecular dynamics is involved. This will not effect the results, but it will improve reproducibility whenever a problem occurs. The random seed should look like the following, where one should change the first number to a desired value:
RANDOM_SEED = 248489752 0 0
Training
We strongly advise to follow this guideline to on-the-fly learning:
- It is generally possible to train force fields on a smaller unit cell and then apply it to a larger system. Be careful to choose the structure large enough, so that the phonons or collective vibrations "fit" into the supercell.
- It is important to learn the correct forces. This requires converged electronic calculations with respect to electronic parameters, i.e., the number of k points, the plane-wave cutoff (ENCUT), electronic minimization algorithm, etc.
- Switch off the symmetry as in molecular-dynamics runs (ISYM=0).
- Lower the integration step (POTIM) if the system contains light elements or increase the mass of the light elements (POMASS) in the INCAR or POTCAR file.
- If possible, heat up the system gradually, i.e., use temperature ramping from a low temperature to a temperature approximately 30% higher than the desired application temperature. The determination of the threshold of the maximum error in forces for Bayesian prediction (ML_CTIFOR) is done on the basis of previous Bayesian error prediction of forces. At the beginning of the on-the-fly learning the force field is very inaccurate and also the prediction of Bayesian errors is inaccurate. In many cases this can lead to an overestimation of ML_CTIFOR. If this happens no further learning will be done since each forthcoming structure has a predicted error that is smaller than the threshold, although not enough training structures and local reference configurations have been collected and the force field has a bad accuracy. The heating of the systems greatly helps to avoid these "unlucky situations", because the error of the system increases with the temperature and the threshold will be easily exceeded, leading to further learning and improvement of the force field. Since the error of the system decreases with decreasing temperature, the threshold would be always stuck at the high temperature errors in cooling runs. Hence we don't advise to use cooling runs for on-the-fly learning. Finally, when using temperature ramps, always be very careful to work in a temperature region where no phase transitions are induced.
- If possible, prefer molecular-dynamics training runs in the NpT ensemble, the additional cell fluctuations improve the robustness of the resulting force field. However, for liquids allow only scaling of the supercell (no change of angles, no individual lattice vector scaling) because otherwise the cell may collapse. This can be achieved with the ICONST file, see 3) and 4) here.
- For simulations without fixed lattice, the plane-wave cutoff ENCUT needs to be set at least 30 percent higher than for fixed volume calculations. Also it is goot to restart often (ML_ISTART=1) to reinitialize the PAW basis of the KS orbitals and avoid Pulay stress.
- When the system contains different components, train them separately first. For instance, when the system has a surface of a crystal and a molecule binding on that surface. First, train the bulk crystal, then the surface, next the isolated molecule, and finally the entire system. This way a significant amount of ab-initio calculation can be saved in the combined system which is anyway the computationally most expensive.
Accuracy
The reachable accuracy of the force fields depends on many factors, e.g., species, temperature, pressure, electronic convergence, machine learning method, etc. In our implementation of the kernel-ridge regression the accuracy of the force fields increases with increasing number of local reference configurations. This increase is below linear and at the same time the computational demand increases linearly. So the user has to trade off between accuracy and efficiency.
Here are some empirical guidelines:
- For a given structure the error increases with increasing temperature and pressure. So the force field should not be trained in conditions that are too far away from the target condition. For example for a production run at 300K it is good to learn above that temperature (500K) to be able to sample more structure that could appear in the production run, but it is not beneficial to learn the same phase at 2000K, since it would most likely lower the accuracy of the force-field.
- Liquids usually need much more training structures and local reference configurations to achieve similar accuracy as solids.
- Typical errors of the force field that should be expected are a few meV/atom for the energies and 100 meV/Angstrom or less for the forces. Errors that are slightly above these values can still be ok, but those calculations should be thoroughly checked for correctness.
Monitoring
The monitoring of your learning can be divided into two parts:
- Molecular-dynamics/ensemble related quantities:
- Monitor your structure visually. This means look at the CONTCAR or XDATCAR files with structure/trajectory viewers. Many times when something goes wrong it can be immediately traced back to unwanted or unphysical deformations.
- Volume and lattice parameters in the OUTCAR and CONTCAR files. It is important to confirm that the average volume stays in the desired region. Strong change of the average volume over time in constant temperature and pressure runs indicates phase transitions or non-properly equilibrated systems.
- Temperature and pressure in the OUTCAR and OSZICAR files. Strong deviations of temperature and pressure with respect to desired ones at the beginning of the calculation indicates non-properly equilibrated systems.
- Pair-correlation functions (PCDAT).
- Machine learning specific quantities in the ML_LOGFILE file:
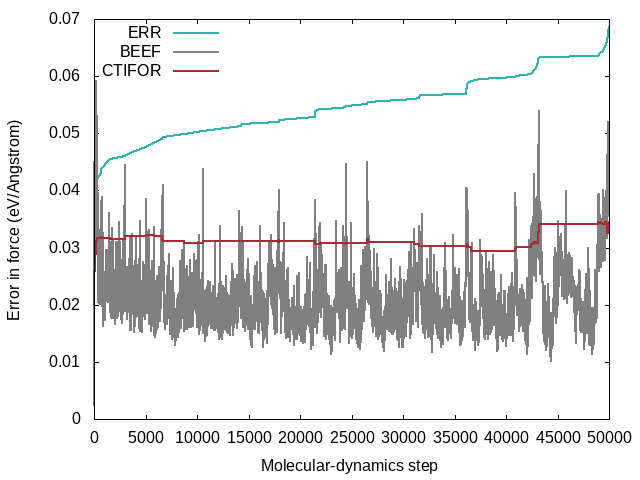
ERR: Root mean square error of predicted energy, forces and stress with respect to ab-initio data for all training structures up to the current molecular-dynamics step.BEEF: Estimated Bayesian error of energy, forces and stress (columns 3-5). Current threshold for the maximum Bayesian error of forces ML_CTIFOR on column 6.THRUPD: Update of ML_CTIFOR.THRHIST: History of Bayesian errors used for ML_CTIFOR.
A typical evolution of the real errors (column 4 of ERR), Bayesian errors (column 4 of BEEF) and threshold (column 6 of BEEF) for the forces looks like the following:
Tuning on-the-fly parameters
In case too many or too few training structures and local reference configurations are selected there are some on-the-fly parameters which can be tuned (for an overview of the learning and threshold algorithms we refer the user to here):
- ML_CTIFOR: Defines the learning threshold for the Bayesian error of the forces for each atom. In a continuation run it can be set to the last value of ML_CTIFOR of the previous run. This way unneccesary sampling at the beginning of the calculation can be skipped. However, when going from one structure to the other, this tag should be very carefully set. ML_CTIFOR is species and system dependent. Low symmetry structures, for example liquids, have usually a much higher error than high symmetry solids for the same compound. If a liquid is learned first and the last ML_CTIFOR from the liquid is used for the corresponding solid, this ML_CTIFOR is way too large for the solid and all predicted errors will be below the threshold. Hence no learning will be done on the solid. In this case it is better to start with the default value for ML_CTIFOR.
- ML_CX: It is involved in the calculation of the threshold, ML_CTIFOR = (average of the stored Bayesian errors in the history) *(1.0 + ML_CX). This tag affects the frequency of selection of training structures and local reference configurations. Positive values of ML_CX result in a less frequent sampling (and hence less ab-initio calculations) and negative values result in the opposite. Typical values of ML_CX are between -0.3 and 0. For training runs using heating, the default usually results in very well balanced machine learned force fields. When the training is performed at a fixed temperature, it is often desirable to decrease to ML_CX=-0.1, in order to increase the number of first principle calculations and thus the size of the training set (the default can result in too few training data).
- ML_HIS: Sets the number of previous Bayesian errors (from learning steps for the default of ML_ICRITERIA) that are used for the update of ML_CTIFOR. If, after the initial phase, strong variations of the Bayesian errors between updates of the threshold appear and the threshold also changes strongly after each update, the default of 10 for this tag can be lowered.
- ML_SCLC_CTIFOR: Scales ML_CTIFOR only in the selection of local reference configurations. In contrast to ML_CX this tag does not affect the frequency of sampling (ab-initio calculations).
- ML_EPS_LOW: Controls the sparsification of the number of local reference configurations after they were selected by the Bayesian error estimation. Increasing ML_EPS_LOW increases the number of local reference configurations that are thrown away and by decreasing the opposite happens. This tag will also not affect the learning frequency since the sparsification is only done after the local reference configurations were selected for a new structure. We do not recommend to increase the threshold to values larger than 1E-7. Below that value this tag works nice to control the number of local reference configurations, however, for multi-component systems the sparsification algorithm tends to lead to strong imbalances in the number of local reference configurations for different species.
- ML_LBASIS_DISCARD: Controls whether the calculation is continued or stopped after the maximum number of local reference configurations ML_MB for any species is reached. The default behaviour (ML_LBASIS_DISCARD=.FALSE.) is that the calculation stops and requests the user to increase ML_MB if that the number of local reference configurations for any species reach ML_MB. In multi-component systems it can happen that, although ML_MB is set to the maximum that is computationally affordable, one species exceeds ML_MB while the other species were not sufficiently sampled and are still far below ML_MB. For that ML_LBASIS_DISCARD=.TRUE. should be set. In that case the fast sampled species would retain the same number of local reference configurations and the new local reference configurations would replace old ones. At the same time new local reference configurations could be added for the slowly-sampled species further improving their accuracy for fitting.
Testing
- Set up an independent test set of random configurations. Then, check the average errors comparing forces, stresses and energy differences of two structures based on DFT and predicted by machine-learned force fields.
- If you have both, ab-initio reference data and a calculation using force fields, check the agreement of some physical properties. For instance, you might check the relaxed lattice parameters, phonons, relative energies of different phases, the elastic constant, the formation of defects etc.
- Plot blocked averages
Application
The following things need to be considered when running only the force field (ML_ISTART=2):
- Set the ab-initio parameters to small values. VASP cannot circumvent the initialization of KS orbitals although they are not used during the molecular-dynamics run with machine learning.
- Monitor the Bayesian error estimates (
BEEF). An increase of this value indicates extrapolation of the force field. In that case the current structure was not contained within the training structures and needs to be included.
Example
Sample input for learning of liquid water in the NpT ensemble at 0.001 kB using a temperature ramp.
ENCUT = 700 #larger cutoff LASPH = .True. GGA = RP IVDW = 11 ALGO = Normal LREAL = Auto ISYM = 0 IBRION = 0 MDALGO = 3 ISIF = 3 POTIM = 1.5 TEBEG = 200 TEEND = 500 LANGEVIN_GAMMA = 10.0 10.0 LANGEVIN_GAMMA_L = 3.0 PMASS = 100 PSTRESS = 0.001 NSW = 20000 POMASS = 8.0 16.0 ML_LMLFF = .TRUE. ML_ISTART = 0
- ENCUT: A larger plane-wave cut off is used to accomodate possible changes of the lattice paramaters, because an NpT ensemble is used (ISIF=3).
- POMASS: Since this structure contains Hydrogen, the mass of Hydrogen is increased by a factor of 8 to be able to use larger integration steps POTIM. Without this one possibly needs to use integration steps of POTIM<0.5 hugely increasing the computation time.
LA 1 2 0 LA 1 3 0 LA 2 3 0 LR 1 0 LR 2 0 LR 3 0 S 1 0 0 0 0 0 0 S 0 1 0 0 0 0 0 S 0 0 1 0 0 0 0 S 0 0 0 1 -1 0 0 S 0 0 0 1 0 -1 0 S 0 0 0 0 1 -1 0
- Since a liquid in the NpT ensemble is simulated here, the ICONST file ensures that the lattice parameters can change to accomodate the pressure, but the length ratio and angles between the lattice parameters is constant. This prevents unwanted deformations of the cell.
Related articles
Machine-learned force fields, Ionic minimization, Molecular dynamics