Liquid Si - MLFF: Difference between revisions
| (24 intermediate revisions by 2 users not shown) | |||
| Line 10: | Line 10: | ||
In this example we start from a 64 atom super cell of diamond-fcc Si (the same as in {{TAG|Liquid Si - Standard MD}}): | In this example we start from a 64 atom super cell of diamond-fcc Si (the same as in {{TAG|Liquid Si - Standard MD}}): | ||
Si_CD_2x2x2 | |||
5.43090000000000 | 5.43090000000000 | ||
2.00000000 0.00000000 0.00000000 | 2.00000000 0.00000000 0.00000000 | ||
| Line 114: | Line 114: | ||
{{TAGBL|NSW}} = 10000 | {{TAGBL|NSW}} = 10000 | ||
{{TAGBL|POTIM}} = 3.0 | {{TAGBL|POTIM}} = 3.0 | ||
{{TAGBL|RANDOM_SEED}} = 88951986 0 0 | |||
#Machine learning paramters | #Machine learning paramters | ||
{{TAGBL| | {{TAGBL|ML_LMLFF}} = .TRUE. | ||
{{TAGBL| | {{TAGBL|ML_ISTART}} = 0 | ||
== Calculation == | == Calculation == | ||
=== Creating the liquid structure === | === Creating the liquid structure === | ||
| Line 165: | Line 129: | ||
After running the calculation, we obtained a force field, but its initial trajectory might be tainted but the unreasonable starting position. Nevertheless, it is instructive to inspect the output to understand how to assess the accuracy of a force field, before refining it in subsequent calculations. | After running the calculation, we obtained a force field, but its initial trajectory might be tainted but the unreasonable starting position. Nevertheless, it is instructive to inspect the output to understand how to assess the accuracy of a force field, before refining it in subsequent calculations. | ||
The main output files for the machine learning are | The main output files for the machine learning are | ||
;{{TAG| | ;{{TAG|ML_ABN}}: contains the ab initio structure datasets used for the learning. It will be needed for continuation runs as {{TAG|ML_AB}}. | ||
;{{TAG| | ;{{TAG|ML_FFN}}: contains the regression results (weights, parameters, etc.). It will be needed for continuation runs as {{TAG|ML_FF}}. | ||
;{{TAG|ML_LOGFILE}}: logging the proceedings of the machine learning. | ;{{TAG|ML_LOGFILE}}: logging the proceedings of the machine learning. This file consists of keywords that are nicely "grepable." The keywords are explained in the in the beginning of the file and upon "grepping". The status of each MD step is given by the keyword "STATUS". Please invoke the following command: | ||
grep STATUS ML_LOGFILE | |||
The output should look similar to the following: | |||
------- | # STATUS ############################################################### | ||
# STATUS This line describes the overall status of each step. | |||
# STATUS | |||
# STATUS nstep ..... MD time step or input structure counter | |||
# STATUS state ..... One-word description of step action | |||
# STATUS - "accurate" (1) : Errors are low, force field is used | |||
# STATUS - "threshold" (2) : Errors exceeded threshold, structure is sampled from ab initio | |||
# STATUS - "learning" (3) : Stored configurations are used for training force field | |||
The | # STATUS - "critical" (4) : Errors are high, ab initio sampling and learning is enforced | ||
# STATUS - "predict" (5) : Force field is used in prediction mode only, no error checking | |||
# STATUS is ........ Integer representation of above one-word description (integer in parenthesis) | |||
# STATUS doabin .... Perform ab initio calculation (T/F) | |||
# STATUS iff ....... Force field available (T/F, False after startup hints to possible convergence problems) | |||
# STATUS nsample ... Number of steps since last reference structure collection (sample = T) | |||
# STATUS ngenff .... Number of steps since last force field generation (genff = T) | |||
# STATUS ############################################################### | |||
# STATUS nstep state is doabin iff nsample ngenff | |||
# STATUS 2 3 4 5 6 7 8 | |||
# STATUS ############################################################### | |||
STATUS 0 threshold 2 T F 0 0 | |||
STATUS 1 critical 4 T F 0 1 | |||
STATUS 2 critical 4 T F 0 2 | |||
STATUS 3 critical 4 T T 0 1 | |||
STATUS 4 critical 4 T T 0 1 | |||
STATUS 5 critical 4 T T 0 1 | |||
. . . . . . . . | |||
. . . . . . . . | |||
. . . . . . . . | |||
STATUS 9997 accurate 1 F T 945 996 | |||
STATUS 9998 accurate 1 F T 946 997 | |||
STATUS 9999 accurate 1 F T 947 998 | |||
STATUS 10000 learning 3 T T 948 999 | |||
Another important keyword is "ERR". For this instance we should type the following command: | |||
grep ERR ML_LOGFILE | |||
The output should look like the following: | |||
# ERR ###################################################################### | |||
# ERR This line contains the RMSEs of the predictions with respect to ab initio results for the training data. | |||
# ERR | |||
# ERR nstep ......... MD time step or input structure counter | |||
# ERR rmse_energy ... RMSE of energies (eV atom^-1) | |||
# ERR rmse_force .... RMSE of forces (eV Angst^-1) | |||
# ERR rmse_stress ... RMSE of stress (kB) | |||
# ERR ###################################################################### | |||
# ERR nstep rmse_energy rmse_force rmse_stress | |||
# ERR 2 3 4 5 | |||
# ERR ###################################################################### | |||
ERR 0 0.00000000E+00 0.00000000E+00 0.00000000E+00 | |||
ERR 1 0.00000000E+00 0.00000000E+00 0.00000000E+00 | |||
ERR 2 0.00000000E+00 0.00000000E+00 0.00000000E+00 | |||
ERR 3 2.84605192E-05 9.82351889E-03 2.40003743E-02 | |||
ERR 4 1.83193349E-05 1.06700600E-02 5.37606479E-02 | |||
ERR 5 4.12132223E-05 1.34123085E-02 1.01588957E-01 | |||
ERR 6 9.51627413E-05 1.90335214E-02 1.31959103E-01 | |||
. . . . . | |||
. . . . . | |||
. . . . . | |||
ERR 9042 1.07159240E-02 2.41283323E-01 4.95695745E+00 | |||
ERR 9052 1.07159240E-02 2.41283323E-01 4.95695745E+00 | |||
ERR 10000 1.07159240E-02 2.41283323E-01 4.95695745E+00 | |||
This tag lists the errors on the energy, forces and stress of the force field compared to the ab initio results on the available training data. The second column shows the MD step. We see that the entry is not output at every MD step. The errors only change if the force field is updated, hence when an ab initio calculation is executed (it should correlate with the doabin column of the STATUS keyword). The other three columns show the errors on the energy (eV/atom), forces (ev/Angstrom) and stress (kB). | |||
=== Structral properties of the force field === | === Structral properties of the force field === | ||
| Line 186: | Line 203: | ||
Now, we proceed with the force field calculation and set up the required files | Now, we proceed with the force field calculation and set up the required files | ||
cp POSCAR.T2000_relaxed {{TAGBL|POSCAR}} | cp POSCAR.T2000_relaxed {{TAGBL|POSCAR}} | ||
cp {{TAGBL| | cp {{TAGBL|ML_FFN}} {{TAGBL|ML_FF}} | ||
To run a shorter simulation using only the force field, we change the following {{TAG|INCAR}} tags to | To run a shorter simulation using only the force field, we change the following {{TAG|INCAR}} tags to | ||
{{TAGBL| | {{TAGBL|ML_ISTART}} = 2 | ||
{{TAGBL|NSW}} = 1000 | {{TAGBL|NSW}} = 1000 | ||
After the calculation finished, we backup the history of the atomic positions | After the calculation finished, we backup the history of the atomic positions | ||
cp {{TAGBL|XDATCAR}} XDATCAR.MLFF_3ps | cp {{TAGBL|XDATCAR}} XDATCAR.MLFF_3ps | ||
To analyze the pair correlation function, we use the PERL script ''pair_correlation_function.pl'' | |||
<div class="toccolours mw-customtoggle-script">'''Click to show/hide pair_correlation_function.pl'''</div> | |||
<div class="mw-collapsible mw-collapsed" id="mw-customcollapsible-script"> | |||
<pre> | |||
#!/usr/bin/perl | |||
use strict; | |||
use warnings; | |||
#configuration for which ensemble average is to be calculated | |||
my $confmin=1; #starting index of configurations in XDATCAR file for pair correlation function | |||
my $confmax=20000; #last index of configurations in XDATCAR file for pair correlation function | |||
my $confskip=1; #stepsize for configuration loop | |||
my $species_1 = 1; #species 1 for which pair correlation function is going to be calculated | |||
my $species_2 = 1; #species 2 for which pair correlation function is going to be calculated | |||
#setting radial grid | |||
my $rmin=0.0; #minimal value of radial grid | |||
my $rmax=10.0; #maximum value of radial grid | |||
my $nr=300; #number of equidistant steps in radial grid | |||
my $dr=($rmax-$rmin)/$nr; #stepsize in radial grid | |||
my $tol=0.0000000001; #tolerance limit for r->0 | |||
my $z=0; #counter | |||
my $numelem; #number of elements | |||
my @elements; #number of atoms per element saved in list/array | |||
my $lattscale; #scaling factor for lattice | |||
my @b; #Bravais matrix | |||
my $nconf=0; #number of configurations in XDATCAR file | |||
my @cart; #Cartesian coordinates for each atom and configuration | |||
my $atmin_1=0; #first index of species one | |||
my $atmax_1; #last index of species one | |||
my $atmin_2=0; #first index of species two | |||
my $atmax_2; #last index of species two | |||
my @vol; #volume of cell (determinant of Bravais matrix) | |||
my $pi=4*atan2(1, 1); #constant pi | |||
my $natom=0; #total number of atoms in cell | |||
my @pcf; #pair correlation function (list/array) | |||
my $mult_x=1; #periodic repetition of cells in x dimension | |||
my $mult_y=1; #periodic repetition of cells in y dimension | |||
my $mult_z=1; #periodic repetition of cells in z dimension | |||
my @cart_super; #Cartesian cells over multiple cells | |||
my @vec_len; #Length of lattice vectors in 3 spatial coordinates | |||
#my $ensemble_type="NpT"; #Set Npt or NVT. Needs to be set since both have different XDATCAR file. | |||
my $ensemble_type="NVT"; #Set Npt or NVT. Needs to be set since both have different XDATCAR file. | |||
my $av_vol=0; #Average volume in cell | |||
#reading in XDATCAR file | |||
while (<>) | |||
{ | |||
chomp; | |||
$_=~s/^/ /; | |||
my @help=split(/[\t,\s]+/); | |||
$z++; | |||
if ($z==2) | |||
{ | |||
$lattscale = $help[1]; | |||
} | |||
if ($z==3) | |||
{ | |||
$b[$nconf+1][1][1]=$help[1]*$lattscale; | |||
$b[$nconf+1][1][2]=$help[2]*$lattscale; | |||
$b[$nconf+1][1][3]=$help[3]*$lattscale; | |||
} | |||
if ($z==4) | |||
{ | |||
$b[$nconf+1][2][1]=$help[1]*$lattscale; | |||
$b[$nconf+1][2][2]=$help[2]*$lattscale; | |||
$b[$nconf+1][2][3]=$help[3]*$lattscale; | |||
} | |||
if ($z==5) | |||
{ | |||
$b[$nconf+1][3][1]=$help[1]*$lattscale; | |||
$b[$nconf+1][3][2]=$help[2]*$lattscale; | |||
$b[$nconf+1][3][3]=$help[3]*$lattscale; | |||
} | |||
if ($z==7) | |||
{ | |||
if ($nconf==0) | |||
{ | |||
$numelem=@help-1; | |||
for (my $i=1;$i<=$numelem;$i++) | |||
{ | |||
$elements[$i]=$help[$i]; | |||
$natom=$natom+$help[$i]; | |||
} | |||
} | |||
} | |||
if ($_=~m/Direct/) | |||
{ | |||
$nconf=$nconf+1; | |||
#for NVT ensemble only one Bravais matrix exists, so it has to be copied | |||
if ($ensemble_type eq "NVT") | |||
{ | |||
for (my $i=1;$i<=3;$i++) | |||
{ | |||
for (my $j=1;$j<=3;$j++) | |||
{ | |||
$b[$nconf][$i][$j]=$b[1][$i][$j]; | |||
} | |||
} | |||
} | |||
for (my $i=1;$i<=$natom;$i++) | |||
{ | |||
$_=<>; | |||
chomp; | |||
$_=~s/^/ /; | |||
my @helpat=split(/[\t,\s]+/); | |||
$cart[$nconf][$i][1]=$b[1][1][1]*$helpat[1]+$b[1][1][2]*$helpat[2]+$b[1][1][3]*$helpat[3]; | |||
$cart[$nconf][$i][2]=$b[1][2][1]*$helpat[1]+$b[1][2][2]*$helpat[2]+$b[1][2][3]*$helpat[3]; | |||
$cart[$nconf][$i][3]=$b[1][3][1]*$helpat[1]+$b[1][3][2]*$helpat[2]+$b[1][3][3]*$helpat[3]; | |||
} | |||
if ($ensemble_type eq "NpT") | |||
{ | |||
$z=0; | |||
} | |||
} | |||
last if eof; | |||
} | |||
if ($confmin>$nconf) | |||
{ | |||
print "Error, confmin larger than number of configurations. Exiting...\n"; | |||
{ | exit; | ||
} | |||
if ($confmax>$nconf) | |||
{ | |||
$confmax=$nconf; | |||
} | |||
for (my $i=1;$i<=$nconf;$i++) | |||
{ | |||
#calculate lattice vector lengths | |||
$vec_len[$i][1]=($b[$i][1][1]*$b[$i][1][1]+$b[$i][1][2]*$b[$i][1][2]+$b[$i][1][3]*$b[$i][1][3])**0.5; | |||
$vec_len[$i][2]=($b[$i][2][1]*$b[$i][2][1]+$b[$i][2][2]*$b[$i][2][2]+$b[$i][2][3]*$b[$i][2][3])**0.5; | |||
$vec_len[$i][3]=($b[$i][3][1]*$b[$i][3][1]+$b[$i][3][2]*$b[$i][3][2]+$b[$i][3][3]*$b[$i][3][3])**0.5; | |||
#calculate volume of cell | |||
$vol[$i]=$b[$i][1][1]*$b[$i][2][2]*$b[$i][3][3]+$b[$i][1][2]*$b[$i][2][3]*$b[$i][3][1]+$b[$i][1][3]*$b[$i][2][1]*$b[$i][3][2]-$b[$i][3][1]*$b[$i][2][2]*$b[$i][1][3]-$b[$i][3][2]*$b[$i][2][3]*$b[$i][1][1]-$b[$i][3][3]*$b[$i][2][1]*$b[$i][1][2]; | |||
$av_vol=$av_vol+$vol[$i]; | |||
} | |||
$av_vol=$av_vol/$nconf; | |||
#choose species 1 for which pair correlation function is going to be calculated | |||
< | $atmin_1=1; | ||
< | if ($species_1>1) | ||
{ | |||
for (my $i=1;$i<$species_1;$i++) | |||
{ | |||
$atmin_1=$atmin_1+$elements[$i]; | |||
} | |||
} | |||
$atmax_1=$atmin_1+$elements[$species_1]-1; | |||
#choose species 2 to which paircorrelation function is calculated to | |||
$atmin_2=1; | |||
if ($species_2>1) | |||
{ | |||
for (my $i=1;$i<$species_2;$i++) | |||
{ | |||
$atmin_2=$atmin_2+$elements[$i]; | |||
} | |||
} | |||
$atmax_2=$atmin_2+$elements[$species_2]-1; | |||
#initialize pair correlation function | |||
for (my $i=0;$i<=($nr-1);$i++) | |||
{ | |||
$pcf[$i]=0.0; | |||
} | |||
# loop over configurations, make histogram of pair correlation function | |||
for (my $j=$confmin;$j<=$confmax;$j=$j+$confskip) | |||
{ | |||
for (my $k=$atmin_1;$k<=$atmax_1;$k++) | |||
{ | |||
for (my $l=$atmin_2;$l<=$atmax_2;$l++) | |||
{ | |||
if ($k==$l) {next}; | |||
for (my $g_x=-$mult_x;$g_x<=$mult_x;$g_x++) | |||
{ | |||
for (my $g_y=-$mult_y;$g_y<=$mult_y;$g_y++) | |||
{ | |||
for (my $g_z=-$mult_y;$g_z<=$mult_z;$g_z++) | |||
{ | |||
my $at2_x=$cart[$j][$l][1]+$vec_len[$j][1]*$g_x; | |||
my $at2_y=$cart[$j][$l][2]+$vec_len[$j][2]*$g_y; | |||
my $at2_z=$cart[$j][$l][3]+$vec_len[$j][3]*$g_z; | |||
my $dist=($cart[$j][$k][1]-$at2_x)**2.0+($cart[$j][$k][2]-$at2_y)**2.0+($cart[$j][$k][3]-$at2_z)**2.0; | |||
$dist=$dist**0.5; | |||
#determine integer multiple | |||
my $zz=int(($dist-$rmin)/$dr+0.5); | |||
if ($zz<$nr) | |||
{ | |||
$pcf[$zz]=$pcf[$zz]+1.0; | |||
} | |||
} | |||
} | |||
} | |||
} | |||
} | |||
} | |||
#make ensemble average, rescale functions and print | |||
for (my $i=0;$i<=($nr-1);$i++) | |||
{ | |||
my $r=$rmin+$i*$dr; | |||
if ($r<$tol) | |||
{ | |||
$pcf[$i]=0.0; | |||
} | |||
else | |||
{ | |||
$pcf[$i]=$pcf[$i]*$av_vol/(4*$pi*$r*$r*$dr*(($confmax-$confmin)/$confskip)*($atmax_2-$atmin_2+1)*($atmax_1-$atmin_1+1));#*((2.0*$mult_x+1.0)*(2.0*$mult_y+1.0)*(2.0*$mult_z+1.0))); | |||
} | |||
print $r," ",$pcf[$i],"\n"; | |||
} | |||
</pre> | |||
</div> | |||
and process the previously saved {{TAG|XDATCAR}} files | and process the previously saved {{TAG|XDATCAR}} files | ||
perl pair_correlation_function.pl XDATCAR.MLFF_3ps > pair_MLFF_3ps.dat | perl pair_correlation_function.pl XDATCAR.MLFF_3ps > pair_MLFF_3ps.dat | ||
To save time the pair correlation function for 1000 ab initio MD steps is precalculated in the file ''pair_AI_3ps.dat''. | |||
The interested user can of course calculate the results of the ab initio MD by rerunning the above steps while switching off machine learning via | |||
{{TAGBL|ML_LMLFF}} = .FALSE. | |||
We can compare the pair correlation functions, e.g. with gnuplot using the following command | We can compare the pair correlation functions, e.g. with gnuplot using the following command | ||
gnuplot -e "set terminal jpeg; set xlabel 'r(Ang)'; set ylabel 'PCF'; set style data lines; plot 'pair_MLFF_3ps.dat', 'pair_AI_3ps.dat' " > PC_MLFF_vs_AI_3ps.jpg | gnuplot -e "set terminal jpeg; set xlabel 'r(Ang)'; set ylabel 'PCF'; set style data lines; plot 'pair_MLFF_3ps.dat', 'pair_AI_3ps.dat' " > PC_MLFF_vs_AI_3ps.jpg | ||
| Line 213: | Line 437: | ||
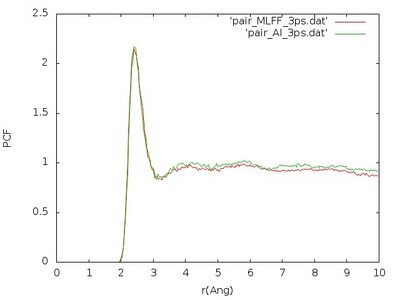
[[File:PC MLFF vs AI 3ps.jpg|400px]] | [[File:PC MLFF vs AI 3ps.jpg|400px]] | ||
We see that pair correlation is quite well reproduced although the error in the force of 0. | We see that pair correlation is quite well reproduced although the error in the force of ~0.242 eV/<math>\AA</math> shown above is a little bit too large. This error is maybe too large for accurate production calculations (usually an accuracy of approximately 0.1 eV/<math>\AA</math> is targeted), but since the pair correlation function is well reproduced it is perfectly fine to use this on-the-fly force field in the time-consuming melting of the crystal. | ||
=== Obtaining a more accurate force field === | === Obtaining a more accurate force field === | ||
| Line 223: | Line 447: | ||
and change the following {{TAG|INCAR}} tags | and change the following {{TAG|INCAR}} tags | ||
{{TAGBL|ALGO}} = Normal | {{TAGBL|ALGO}} = Normal | ||
{{TAGBL| | {{TAGBL|ML_LMLFF}} = .TRUE. | ||
{{TAGBL| | {{TAGBL|ML_ISTART}} = 0 | ||
{{TAGBL|NSW}} = | {{TAGBL|NSW}} = 1000 | ||
If you run have resources to run in parallel, you can reduce the computation time further by adding k point parallelization with the {{TAG|KPAR}} tag. | If you run have resources to run in parallel, you can reduce the computation time further by adding k point parallelization with the {{TAG|KPAR}} tag. | ||
We use a denser k-point mesh in the {{TAG|KPOINTS}} file | We use a denser k-point mesh in the {{TAG|KPOINTS}} file | ||
| Line 233: | Line 457: | ||
2 2 2 | 2 2 2 | ||
0 0 0 | 0 0 0 | ||
We will learn a new force field | We will learn a new force field with 1000 MD steps (each of 3 fs). Keep in mind to run the calculation using the '''standard''' version of VASP (usually ''vasp_std''). | ||
After running the calculation, we examine the error in the {{TAG|ML_LOGFILE}} | After running the calculation, we examine the error "ERR" in the {{TAG|ML_LOGFILE}} by typing: | ||
grep "ERR" ML_LOGFILE | |||
where the last entries should be close to | |||
---- | ERR 886 5.98467749E-03 1.48190308E-01 2.38264786E+00 | ||
ERR 908 5.98467749E-03 1.48190308E-01 2.38264786E+00 | |||
ERR 925 5.98467749E-03 1.48190308E-01 2.38264786E+00 | |||
ERR 959 5.98467749E-03 1.48190308E-01 2.38264786E+00 | |||
ERR 980 5.98467749E-03 1.48190308E-01 2.38264786E+00 | |||
ERR 990 5.99559653E-03 1.50261779E-01 2.40349561E+00 | |||
ERR 1000 5.99559653E-03 1.50261779E-01 2.40349561E+00 | |||
We immediately see that the errors are significantly lower than in the previous calculation with only one k point. This is due to the less noisy ab initio data which is easier to learn. | We immediately see that the errors for the forces are significantly lower than in the previous calculation with only one k point. This is due to the less noisy ab initio data which is easier to learn. | ||
To understand how the force field is learned, we inspect the {{TAG| | To understand how the force field is learned, we inspect the {{TAG|ML_ABN}} file containing the training data. In the beginning of this file, you will find information about the number of reference structures for training | ||
1.0 Version | |||
************************************************** | ************************************************** | ||
The number of configurations | The number of configurations | ||
-------------------------------------------------- | -------------------------------------------------- | ||
48 | |||
and the size of the basis set | and the number of local reference configurations (size of the basis set) | ||
************************************************** | ************************************************** | ||
The numbers of basis sets per atom type | The numbers of basis sets per atom type | ||
-------------------------------------------------- | -------------------------------------------------- | ||
382 | |||
We will | We will further continue the training a 1000 MD steps and see how the number of training structures and the number of local reference configurations change. | ||
cp {{TAGBL| | Do the following steps: | ||
cp {{TAGBL|ML_ABN}} {{TAGBL|ML_AB}} | |||
cp {{TAGBL|CONTCAR}} {{TAGBL|POSCAR}} | cp {{TAGBL|CONTCAR}} {{TAGBL|POSCAR}} | ||
and set the following {{TAG|INCAR}} | and set the following {{TAG|INCAR}} tag | ||
{{TAGBL| | {{TAGBL|ML_ISTART}} = 1 | ||
After running the calculation, we inspect the last instance of the errors in the {{TAG|ML_LOGFILE}} | After running the calculation, we inspect the last instance of the errors in the {{TAG|ML_LOGFILE}} by typing: | ||
grep ERR ML_LOGFILE | |||
The last few lines should have values close to: | |||
-- | ERR 675 5.10937061E-03 1.46895065E-01 2.50094941E+00 | ||
ERR 861 5.10937061E-03 1.46895065E-01 2.50094941E+00 | |||
ERR 924 5.10937061E-03 1.46895065E-01 2.50094941E+00 | |||
ERR 942 5.01460989E-03 1.47816836E-01 2.47329693E+00 | |||
ERR 1000 5.01460989E-03 1.47816836E-01 2.47329693E+00 | |||
We see that the accuracy has changed slightly. We also look at the ML_ABN file and the number of reference structures for training should increase compared to the run before | |||
1.0 Version | |||
We see that the accuracy has | |||
************************************************** | ************************************************** | ||
The number of configurations | The number of configurations | ||
-------------------------------------------------- | -------------------------------------------------- | ||
99 | |||
Also the number of local reference configurations (basis sets) increases compared to the previous calculation | |||
************************************************** | ************************************************** | ||
The numbers of basis sets per atom type | The numbers of basis sets per atom type | ||
-------------------------------------------------- | -------------------------------------------------- | ||
665 | |||
Ideally, one should continue learning until no structures need to be added to the training data and basis set anymore. Very often this can take up to 100ps depending on the material and conditions. In practice, the prediction of the Bayesian error exhibits numerical inaccuracies so that an ab initio calculation is conducted from time to time even if the force field is accurate enough. So measuring only the decreasing frequency of addition of new data is not sufficient to know when to finish. One should also look at the accuracy of the force on the training data and more importantly on the accuracy on some test data that is outside of the training sets. | |||
== Download == | == Download == | ||
[[Media: | [[Media:MLFF_Liquid_Si_tutorial.tgz| MLFF_Liquid_Si_tutorial.tgz]] | ||
{{Template:Machine learning force field - Tutorial}} | {{Template:Machine learning force field - Tutorial}} | ||
[[Category:Examples]] | [[Category:Examples]] | ||
Latest revision as of 15:13, 15 April 2022
Task
Generating a machine learning force field for liquid Si. For this tutorial, we expect that the user is already familiar with running conventional ab initio molecular dynamic calculations.
Input
POSCAR
In this example we start from a 64 atom super cell of diamond-fcc Si (the same as in Liquid Si - Standard MD):
Si_CD_2x2x2
5.43090000000000
2.00000000 0.00000000 0.00000000
0.00000000 2.00000000 0.00000000
0.00000000 0.00000000 2.00000000
Si
64
Direct
0.00000000 0.00000000 0.00000000
0.50000000 0.00000000 0.00000000
0.00000000 0.50000000 0.00000000
0.50000000 0.50000000 0.00000000
0.00000000 0.00000000 0.50000000
0.50000000 0.00000000 0.50000000
0.00000000 0.50000000 0.50000000
0.50000000 0.50000000 0.50000000
0.37500000 0.12500000 0.37500000
0.87500000 0.12500000 0.37500000
0.37500000 0.62500000 0.37500000
0.87500000 0.62500000 0.37500000
0.37500000 0.12500000 0.87500000
0.87500000 0.12500000 0.87500000
0.37500000 0.62500000 0.87500000
0.87500000 0.62500000 0.87500000
0.00000000 0.25000000 0.25000000
0.50000000 0.25000000 0.25000000
0.00000000 0.75000000 0.25000000
0.50000000 0.75000000 0.25000000
0.00000000 0.25000000 0.75000000
0.50000000 0.25000000 0.75000000
0.00000000 0.75000000 0.75000000
0.50000000 0.75000000 0.75000000
0.37500000 0.37500000 0.12500000
0.87500000 0.37500000 0.12500000
0.37500000 0.87500000 0.12500000
0.87500000 0.87500000 0.12500000
0.37500000 0.37500000 0.62500000
0.87500000 0.37500000 0.62500000
0.37500000 0.87500000 0.62500000
0.87500000 0.87500000 0.62500000
0.25000000 0.00000000 0.25000000
0.75000000 0.00000000 0.25000000
0.25000000 0.50000000 0.25000000
0.75000000 0.50000000 0.25000000
0.25000000 0.00000000 0.75000000
0.75000000 0.00000000 0.75000000
0.25000000 0.50000000 0.75000000
0.75000000 0.50000000 0.75000000
0.12500000 0.12500000 0.12500000
0.62500000 0.12500000 0.12500000
0.12500000 0.62500000 0.12500000
0.62500000 0.62500000 0.12500000
0.12500000 0.12500000 0.62500000
0.62500000 0.12500000 0.62500000
0.12500000 0.62500000 0.62500000
0.62500000 0.62500000 0.62500000
0.25000000 0.25000000 0.00000000
0.75000000 0.25000000 0.00000000
0.25000000 0.75000000 0.00000000
0.75000000 0.75000000 0.00000000
0.25000000 0.25000000 0.50000000
0.75000000 0.25000000 0.50000000
0.25000000 0.75000000 0.50000000
0.75000000 0.75000000 0.50000000
0.12500000 0.37500000 0.37500000
0.62500000 0.37500000 0.37500000
0.12500000 0.87500000 0.37500000
0.62500000 0.87500000 0.37500000
0.12500000 0.37500000 0.87500000
0.62500000 0.37500000 0.87500000
0.12500000 0.87500000 0.87500000
0.62500000 0.87500000 0.87500000
KPOINTS
We will start with a single k point in this example:
K-Points 0 Gamma 1 1 1 0 0 0
INCAR
#Basic parameters ISMEAR = 0 SIGMA = 0.1 LREAL = Auto ISYM = -1 NELM = 100 EDIFF = 1E-4 LWAVE = .FALSE. LCHARG = .FALSE. #Parallelization of ab initio calculations NCORE = 2 #MD IBRION = 0 MDALGO = 2 ISIF = 2 SMASS = 1.0 TEBEG = 2000 NSW = 10000 POTIM = 3.0 RANDOM_SEED = 88951986 0 0 #Machine learning paramters ML_LMLFF = .TRUE. ML_ISTART = 0
Calculation
Creating the liquid structure
Because we don't have a structure of liquid silicon readily available, we first create that structure by starting from a super cell of crystalline silicon with 64 atoms. The temperature is set to 2000 K so that the crystal melts rapidly in the MD run. To improve the simulation speed drastically, we utilize the on-the-fly machine learning. Most of the ab initio steps will be replaced by very fast force-field ones. Within 10000 steps equivalent to 30 ps, we have obtained a good starting position for the subsequent simulations in the CONTCAR file. You can copy the input files or download them.
After running the calculation, we obtained a force field, but its initial trajectory might be tainted but the unreasonable starting position. Nevertheless, it is instructive to inspect the output to understand how to assess the accuracy of a force field, before refining it in subsequent calculations. The main output files for the machine learning are
- ML_ABN
- contains the ab initio structure datasets used for the learning. It will be needed for continuation runs as ML_AB.
- ML_FFN
- contains the regression results (weights, parameters, etc.). It will be needed for continuation runs as ML_FF.
- ML_LOGFILE
- logging the proceedings of the machine learning. This file consists of keywords that are nicely "grepable." The keywords are explained in the in the beginning of the file and upon "grepping". The status of each MD step is given by the keyword "STATUS". Please invoke the following command:
grep STATUS ML_LOGFILE
The output should look similar to the following:
# STATUS ###############################################################
# STATUS This line describes the overall status of each step.
# STATUS
# STATUS nstep ..... MD time step or input structure counter
# STATUS state ..... One-word description of step action
# STATUS - "accurate" (1) : Errors are low, force field is used
# STATUS - "threshold" (2) : Errors exceeded threshold, structure is sampled from ab initio
# STATUS - "learning" (3) : Stored configurations are used for training force field
# STATUS - "critical" (4) : Errors are high, ab initio sampling and learning is enforced
# STATUS - "predict" (5) : Force field is used in prediction mode only, no error checking
# STATUS is ........ Integer representation of above one-word description (integer in parenthesis)
# STATUS doabin .... Perform ab initio calculation (T/F)
# STATUS iff ....... Force field available (T/F, False after startup hints to possible convergence problems)
# STATUS nsample ... Number of steps since last reference structure collection (sample = T)
# STATUS ngenff .... Number of steps since last force field generation (genff = T)
# STATUS ###############################################################
# STATUS nstep state is doabin iff nsample ngenff
# STATUS 2 3 4 5 6 7 8
# STATUS ###############################################################
STATUS 0 threshold 2 T F 0 0
STATUS 1 critical 4 T F 0 1
STATUS 2 critical 4 T F 0 2
STATUS 3 critical 4 T T 0 1
STATUS 4 critical 4 T T 0 1
STATUS 5 critical 4 T T 0 1
. . . . . . . .
. . . . . . . .
. . . . . . . .
STATUS 9997 accurate 1 F T 945 996
STATUS 9998 accurate 1 F T 946 997
STATUS 9999 accurate 1 F T 947 998
STATUS 10000 learning 3 T T 948 999
Another important keyword is "ERR". For this instance we should type the following command:
grep ERR ML_LOGFILE
The output should look like the following:
# ERR ###################################################################### # ERR This line contains the RMSEs of the predictions with respect to ab initio results for the training data. # ERR # ERR nstep ......... MD time step or input structure counter # ERR rmse_energy ... RMSE of energies (eV atom^-1) # ERR rmse_force .... RMSE of forces (eV Angst^-1) # ERR rmse_stress ... RMSE of stress (kB) # ERR ###################################################################### # ERR nstep rmse_energy rmse_force rmse_stress # ERR 2 3 4 5 # ERR ###################################################################### ERR 0 0.00000000E+00 0.00000000E+00 0.00000000E+00 ERR 1 0.00000000E+00 0.00000000E+00 0.00000000E+00 ERR 2 0.00000000E+00 0.00000000E+00 0.00000000E+00 ERR 3 2.84605192E-05 9.82351889E-03 2.40003743E-02 ERR 4 1.83193349E-05 1.06700600E-02 5.37606479E-02 ERR 5 4.12132223E-05 1.34123085E-02 1.01588957E-01 ERR 6 9.51627413E-05 1.90335214E-02 1.31959103E-01 . . . . . . . . . . . . . . . ERR 9042 1.07159240E-02 2.41283323E-01 4.95695745E+00 ERR 9052 1.07159240E-02 2.41283323E-01 4.95695745E+00 ERR 10000 1.07159240E-02 2.41283323E-01 4.95695745E+00
This tag lists the errors on the energy, forces and stress of the force field compared to the ab initio results on the available training data. The second column shows the MD step. We see that the entry is not output at every MD step. The errors only change if the force field is updated, hence when an ab initio calculation is executed (it should correlate with the doabin column of the STATUS keyword). The other three columns show the errors on the energy (eV/atom), forces (ev/Angstrom) and stress (kB).
Structral properties of the force field
To examine the accuracy of structural properties, we compare the deviations between a 3 ps molecular dynamics run using the force field and a full ab initio calculation. For a meaningful comparison, it is best to start from the same initial structure. We will use the liquid structure, we obtained in the previous step and back it up
cp CONTCAR POSCAR.T2000_relaxed
Now, we proceed with the force field calculation and set up the required files
cp POSCAR.T2000_relaxed POSCAR cp ML_FFN ML_FF
To run a shorter simulation using only the force field, we change the following INCAR tags to
ML_ISTART = 2 NSW = 1000
After the calculation finished, we backup the history of the atomic positions
cp XDATCAR XDATCAR.MLFF_3ps
To analyze the pair correlation function, we use the PERL script pair_correlation_function.pl
#!/usr/bin/perl
use strict;
use warnings;
#configuration for which ensemble average is to be calculated
my $confmin=1; #starting index of configurations in XDATCAR file for pair correlation function
my $confmax=20000; #last index of configurations in XDATCAR file for pair correlation function
my $confskip=1; #stepsize for configuration loop
my $species_1 = 1; #species 1 for which pair correlation function is going to be calculated
my $species_2 = 1; #species 2 for which pair correlation function is going to be calculated
#setting radial grid
my $rmin=0.0; #minimal value of radial grid
my $rmax=10.0; #maximum value of radial grid
my $nr=300; #number of equidistant steps in radial grid
my $dr=($rmax-$rmin)/$nr; #stepsize in radial grid
my $tol=0.0000000001; #tolerance limit for r->0
my $z=0; #counter
my $numelem; #number of elements
my @elements; #number of atoms per element saved in list/array
my $lattscale; #scaling factor for lattice
my @b; #Bravais matrix
my $nconf=0; #number of configurations in XDATCAR file
my @cart; #Cartesian coordinates for each atom and configuration
my $atmin_1=0; #first index of species one
my $atmax_1; #last index of species one
my $atmin_2=0; #first index of species two
my $atmax_2; #last index of species two
my @vol; #volume of cell (determinant of Bravais matrix)
my $pi=4*atan2(1, 1); #constant pi
my $natom=0; #total number of atoms in cell
my @pcf; #pair correlation function (list/array)
my $mult_x=1; #periodic repetition of cells in x dimension
my $mult_y=1; #periodic repetition of cells in y dimension
my $mult_z=1; #periodic repetition of cells in z dimension
my @cart_super; #Cartesian cells over multiple cells
my @vec_len; #Length of lattice vectors in 3 spatial coordinates
#my $ensemble_type="NpT"; #Set Npt or NVT. Needs to be set since both have different XDATCAR file.
my $ensemble_type="NVT"; #Set Npt or NVT. Needs to be set since both have different XDATCAR file.
my $av_vol=0; #Average volume in cell
#reading in XDATCAR file
while (<>)
{
chomp;
$_=~s/^/ /;
my @help=split(/[\t,\s]+/);
$z++;
if ($z==2)
{
$lattscale = $help[1];
}
if ($z==3)
{
$b[$nconf+1][1][1]=$help[1]*$lattscale;
$b[$nconf+1][1][2]=$help[2]*$lattscale;
$b[$nconf+1][1][3]=$help[3]*$lattscale;
}
if ($z==4)
{
$b[$nconf+1][2][1]=$help[1]*$lattscale;
$b[$nconf+1][2][2]=$help[2]*$lattscale;
$b[$nconf+1][2][3]=$help[3]*$lattscale;
}
if ($z==5)
{
$b[$nconf+1][3][1]=$help[1]*$lattscale;
$b[$nconf+1][3][2]=$help[2]*$lattscale;
$b[$nconf+1][3][3]=$help[3]*$lattscale;
}
if ($z==7)
{
if ($nconf==0)
{
$numelem=@help-1;
for (my $i=1;$i<=$numelem;$i++)
{
$elements[$i]=$help[$i];
$natom=$natom+$help[$i];
}
}
}
if ($_=~m/Direct/)
{
$nconf=$nconf+1;
#for NVT ensemble only one Bravais matrix exists, so it has to be copied
if ($ensemble_type eq "NVT")
{
for (my $i=1;$i<=3;$i++)
{
for (my $j=1;$j<=3;$j++)
{
$b[$nconf][$i][$j]=$b[1][$i][$j];
}
}
}
for (my $i=1;$i<=$natom;$i++)
{
$_=<>;
chomp;
$_=~s/^/ /;
my @helpat=split(/[\t,\s]+/);
$cart[$nconf][$i][1]=$b[1][1][1]*$helpat[1]+$b[1][1][2]*$helpat[2]+$b[1][1][3]*$helpat[3];
$cart[$nconf][$i][2]=$b[1][2][1]*$helpat[1]+$b[1][2][2]*$helpat[2]+$b[1][2][3]*$helpat[3];
$cart[$nconf][$i][3]=$b[1][3][1]*$helpat[1]+$b[1][3][2]*$helpat[2]+$b[1][3][3]*$helpat[3];
}
if ($ensemble_type eq "NpT")
{
$z=0;
}
}
last if eof;
}
if ($confmin>$nconf)
{
print "Error, confmin larger than number of configurations. Exiting...\n";
exit;
}
if ($confmax>$nconf)
{
$confmax=$nconf;
}
for (my $i=1;$i<=$nconf;$i++)
{
#calculate lattice vector lengths
$vec_len[$i][1]=($b[$i][1][1]*$b[$i][1][1]+$b[$i][1][2]*$b[$i][1][2]+$b[$i][1][3]*$b[$i][1][3])**0.5;
$vec_len[$i][2]=($b[$i][2][1]*$b[$i][2][1]+$b[$i][2][2]*$b[$i][2][2]+$b[$i][2][3]*$b[$i][2][3])**0.5;
$vec_len[$i][3]=($b[$i][3][1]*$b[$i][3][1]+$b[$i][3][2]*$b[$i][3][2]+$b[$i][3][3]*$b[$i][3][3])**0.5;
#calculate volume of cell
$vol[$i]=$b[$i][1][1]*$b[$i][2][2]*$b[$i][3][3]+$b[$i][1][2]*$b[$i][2][3]*$b[$i][3][1]+$b[$i][1][3]*$b[$i][2][1]*$b[$i][3][2]-$b[$i][3][1]*$b[$i][2][2]*$b[$i][1][3]-$b[$i][3][2]*$b[$i][2][3]*$b[$i][1][1]-$b[$i][3][3]*$b[$i][2][1]*$b[$i][1][2];
$av_vol=$av_vol+$vol[$i];
}
$av_vol=$av_vol/$nconf;
#choose species 1 for which pair correlation function is going to be calculated
$atmin_1=1;
if ($species_1>1)
{
for (my $i=1;$i<$species_1;$i++)
{
$atmin_1=$atmin_1+$elements[$i];
}
}
$atmax_1=$atmin_1+$elements[$species_1]-1;
#choose species 2 to which paircorrelation function is calculated to
$atmin_2=1;
if ($species_2>1)
{
for (my $i=1;$i<$species_2;$i++)
{
$atmin_2=$atmin_2+$elements[$i];
}
}
$atmax_2=$atmin_2+$elements[$species_2]-1;
#initialize pair correlation function
for (my $i=0;$i<=($nr-1);$i++)
{
$pcf[$i]=0.0;
}
# loop over configurations, make histogram of pair correlation function
for (my $j=$confmin;$j<=$confmax;$j=$j+$confskip)
{
for (my $k=$atmin_1;$k<=$atmax_1;$k++)
{
for (my $l=$atmin_2;$l<=$atmax_2;$l++)
{
if ($k==$l) {next};
for (my $g_x=-$mult_x;$g_x<=$mult_x;$g_x++)
{
for (my $g_y=-$mult_y;$g_y<=$mult_y;$g_y++)
{
for (my $g_z=-$mult_y;$g_z<=$mult_z;$g_z++)
{
my $at2_x=$cart[$j][$l][1]+$vec_len[$j][1]*$g_x;
my $at2_y=$cart[$j][$l][2]+$vec_len[$j][2]*$g_y;
my $at2_z=$cart[$j][$l][3]+$vec_len[$j][3]*$g_z;
my $dist=($cart[$j][$k][1]-$at2_x)**2.0+($cart[$j][$k][2]-$at2_y)**2.0+($cart[$j][$k][3]-$at2_z)**2.0;
$dist=$dist**0.5;
#determine integer multiple
my $zz=int(($dist-$rmin)/$dr+0.5);
if ($zz<$nr)
{
$pcf[$zz]=$pcf[$zz]+1.0;
}
}
}
}
}
}
}
#make ensemble average, rescale functions and print
for (my $i=0;$i<=($nr-1);$i++)
{
my $r=$rmin+$i*$dr;
if ($r<$tol)
{
$pcf[$i]=0.0;
}
else
{
$pcf[$i]=$pcf[$i]*$av_vol/(4*$pi*$r*$r*$dr*(($confmax-$confmin)/$confskip)*($atmax_2-$atmin_2+1)*($atmax_1-$atmin_1+1));#*((2.0*$mult_x+1.0)*(2.0*$mult_y+1.0)*(2.0*$mult_z+1.0)));
}
print $r," ",$pcf[$i],"\n";
}
and process the previously saved XDATCAR files
perl pair_correlation_function.pl XDATCAR.MLFF_3ps > pair_MLFF_3ps.dat
To save time the pair correlation function for 1000 ab initio MD steps is precalculated in the file pair_AI_3ps.dat.
The interested user can of course calculate the results of the ab initio MD by rerunning the above steps while switching off machine learning via
ML_LMLFF = .FALSE.
We can compare the pair correlation functions, e.g. with gnuplot using the following command
gnuplot -e "set terminal jpeg; set xlabel 'r(Ang)'; set ylabel 'PCF'; set style data lines; plot 'pair_MLFF_3ps.dat', 'pair_AI_3ps.dat' " > PC_MLFF_vs_AI_3ps.jpg
The pair correlation functions obtained that way should look similar to this figure
We see that pair correlation is quite well reproduced although the error in the force of ~0.242 eV/ shown above is a little bit too large. This error is maybe too large for accurate production calculations (usually an accuracy of approximately 0.1 eV/ is targeted), but since the pair correlation function is well reproduced it is perfectly fine to use this on-the-fly force field in the time-consuming melting of the crystal.
Obtaining a more accurate force field
Including the melting phase in the force field may impact the accuracy of the force field. To improve it is usually advisable to learn on the pure structures, which in our case this means to use the CONTCAR file obtained after the melting. Furthermore, the force field was learned using only a single k point so that we can improve the accuracy of the reference data by including more k points. In most calculations, it is important to conduct accurate ab initio calculations since bad reference data can limit the accuracy or even inhibit the learning of a force field.
We restart from the liquid structure obtained before
cp POSCAR.T2000_relaxed POSCAR
and change the following INCAR tags
ALGO = Normal ML_LMLFF = .TRUE. ML_ISTART = 0 NSW = 1000
If you run have resources to run in parallel, you can reduce the computation time further by adding k point parallelization with the KPAR tag. We use a denser k-point mesh in the KPOINTS file
2x2x2 Gamma-centered mesh 0 0 0 Gamma 2 2 2 0 0 0
We will learn a new force field with 1000 MD steps (each of 3 fs). Keep in mind to run the calculation using the standard version of VASP (usually vasp_std). After running the calculation, we examine the error "ERR" in the ML_LOGFILE by typing:
grep "ERR" ML_LOGFILE
where the last entries should be close to
ERR 886 5.98467749E-03 1.48190308E-01 2.38264786E+00 ERR 908 5.98467749E-03 1.48190308E-01 2.38264786E+00 ERR 925 5.98467749E-03 1.48190308E-01 2.38264786E+00 ERR 959 5.98467749E-03 1.48190308E-01 2.38264786E+00 ERR 980 5.98467749E-03 1.48190308E-01 2.38264786E+00 ERR 990 5.99559653E-03 1.50261779E-01 2.40349561E+00 ERR 1000 5.99559653E-03 1.50261779E-01 2.40349561E+00
We immediately see that the errors for the forces are significantly lower than in the previous calculation with only one k point. This is due to the less noisy ab initio data which is easier to learn.
To understand how the force field is learned, we inspect the ML_ABN file containing the training data. In the beginning of this file, you will find information about the number of reference structures for training
1.0 Version
**************************************************
The number of configurations
--------------------------------------------------
48
and the number of local reference configurations (size of the basis set)
**************************************************
The numbers of basis sets per atom type
--------------------------------------------------
382
We will further continue the training a 1000 MD steps and see how the number of training structures and the number of local reference configurations change. Do the following steps:
cp ML_ABN ML_AB cp CONTCAR POSCAR
and set the following INCAR tag
ML_ISTART = 1
After running the calculation, we inspect the last instance of the errors in the ML_LOGFILE by typing:
grep ERR ML_LOGFILE
The last few lines should have values close to:
ERR 675 5.10937061E-03 1.46895065E-01 2.50094941E+00 ERR 861 5.10937061E-03 1.46895065E-01 2.50094941E+00 ERR 924 5.10937061E-03 1.46895065E-01 2.50094941E+00 ERR 942 5.01460989E-03 1.47816836E-01 2.47329693E+00 ERR 1000 5.01460989E-03 1.47816836E-01 2.47329693E+00
We see that the accuracy has changed slightly. We also look at the ML_ABN file and the number of reference structures for training should increase compared to the run before
1.0 Version
**************************************************
The number of configurations
--------------------------------------------------
99
Also the number of local reference configurations (basis sets) increases compared to the previous calculation
**************************************************
The numbers of basis sets per atom type
--------------------------------------------------
665
Ideally, one should continue learning until no structures need to be added to the training data and basis set anymore. Very often this can take up to 100ps depending on the material and conditions. In practice, the prediction of the Bayesian error exhibits numerical inaccuracies so that an ab initio calculation is conducted from time to time even if the force field is accurate enough. So measuring only the decreasing frequency of addition of new data is not sufficient to know when to finish. One should also look at the accuracy of the force on the training data and more importantly on the accuracy on some test data that is outside of the training sets.